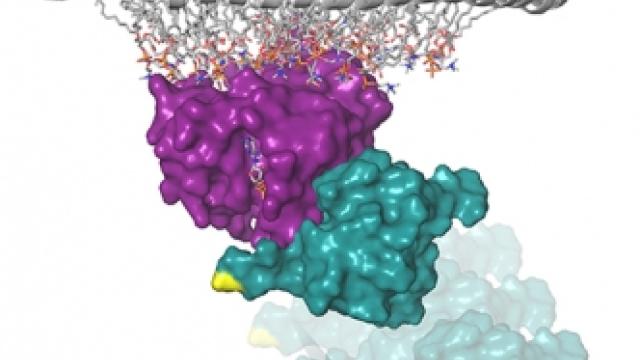
Within families of Ras/Raf proteins, individual preferences matter.

The Ras and Raf family members of proteins are depicted as locks and keys, respectively, to demonstrate how they interact with each other. Morrison and her team suggest that C-Raf is the skeleton key that binds all Ras family members with high affinity, whereas B-Raf exhibits a striking preference for K-Ras. The study helps explain why K-Ras is the most commonly mutated Ras gene in human tumors and why certain B-Raf mutations often co-occur with H-Ras mutations in specific cancers. The findings suggest that Ras/Raf binding preferences need to be considered when treating patients.
Credit: Joseph Meyer, Scientific Publications, Graphics and Media, Frederick National Laboratory, NCI, NIH
Some of the most notorious proteins in cancer biology belong to a family of growth-promoting enzymes known as Ras. These proteins are essential for normal development, but when they become too active, they can drive excessive cell growth. Mutations in the genes that encode Ras proteins can send cells’ growth-signaling pathways into overdrive and are thought to fuel more than 30 percent of human cancers.
Deborah Morrison, Ph.D., has long focused on understanding exactly how Ras proteins cooperate with signaling partners, known as Raf proteins, to drive cell growth. The fundamentals are well known: Ras proteins activate Raf proteins, which in turn trigger a cascade of signaling events that spur cell growth. But human cells have three different Ras proteins — H-Ras, K-Ras and N-Ras — and three different Raf proteins — A-Raf, B-Raf and C-Raf — and researchers have struggled to clarify each family member’s particular role.
Morrison and Research Biologist Elizabeth Terrell, M.S., have now worked out exactly which of these proteins are most likely to interact with one another inside living cells. Their findings, reported in Molecular Cell, have implications for the treatment of patients whose cancers carry Ras or Raf gene mutations, as well as for understanding a group of rare developmental disorders caused by Ras gene mutations.
Until now, scientists have mostly used fragments of Ras and Raf proteins to study how these molecules interact. Morrison’s team turned to a technique called bioluminescence resonance energy transfer (BRET) to instead monitor intact proteins’ associations with one another inside living cells. Using BRET, which indicates when two proteins are in close proximity, the researchers were able to map out the binding preferences of each of the Ras and Raf proteins.
They found that B-Raf, which is the most potent activator of cell growth in the Raf family, binds readily to K-Ras but is much less likely to associate with other members of the Ras family. K-Ras, on the other hand, is able to bind to all of the Raf family members with high affinity. Morrison says these findings help provide an explanation for why K-Ras is the most commonly mutated Ras gene in human tumors and why certain B-Raf mutations often co-occur with H-Ras mutations in specific cancers.
The findings also shed light on how patients with mutations in certain Ras or Raf genes may respond to targeted therapies and suggests that Ras/Raf binding dynamics must be considered when treating patients. Binding preferences may explain, for example, why patients with melanoma have been found to be at risk of developing secondary, H-Ras-driven cancers if they are treated with drugs that inhibit B-Raf.
Finally, Morrison says her team’s experiments demonstrated the power of BRET, not just to assess Ras/Raf interactions, but also to identify factors that either strengthen or interrupt them. They have begun using the method to search for small molecules that inhibit these interactions, with an eye toward developing new cancer treatments.