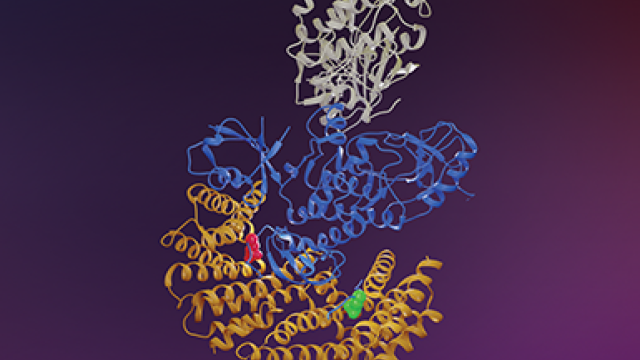
The Ras and Raf family members are depicted as locks and keys, respectively. Morrison, et al., suggest that C-Raf is the skeleton key that binds all Ras family members with high affinity, whereas B-Raf exhibits a striking preference for K-Ras.
Photo credit: Joseph Meyer, Scientific Publications, Graphics and Media, Frederick National Laboratory, NCI, NIH
Using living cells, researchers in CCR have found that a set of enzymatic proteins, known as Raf kinases, have differing affinities in how they bind to a class of cancer-related proteins, known as the Ras GTPases. The findings, from Deborah K. Morrison, Ph.D., Chief of the Laboratory of Cell and Developmental Signaling, and colleagues, appeared December 19, 2019, in Molecular Cell.
Human cells have three different Raf kinases (A, B and C-Raf), and three different Ras members (H, K and N-Ras), and scientists have known that Ras regulates Raf activity to send signals that promote cell growth. Alterations in Ras signaling can lead to disease, including cancer and certain developmental disorders, when genes encoding Ras or Raf proteins are mutated. Most significantly, disease-related mutations can lock Ras into a constitutively active state resulting in continuous cell growth and tumor formation.
Using a technique known as bioluminescence resonance energy transfer (BRET), Morrison and colleagues determined the ability of individual Raf enzymes and Ras proteins to interact with one another in laboratory cultures of live human cells. Most prior research studying Ras and Raf binding had used purified proteins or samples extracted from cells, thereby limiting the understanding of this complex biological interaction. In this new study, the scientists uncovered a set of specific binding preferences that could impact cancer progression and potentially alter how a cancer cell responds to targeted therapies.
The investigators were able to pinpoint specific aspects of Raf and Ras binding that were unique, defining regions in each of the proteins that are responsible for the observed binding preferences. Most notably, they found that B-Raf strongly preferred binding to K-Ras over other Ras family members. This binding pair, K-Ras and B-Raf, also represents the most frequently mutated members of their respective families.
“It turns out that the binding preferences of two major cancer-associated protein families may impact how we view and treat certain tumors and malignancies. Our study also provides the cautionary tale that certain drug treatments can alter Ras/Raf binding dynamics to promote Ras-driven cancer progression,” said Morrison. “Specifically, this finding may explain why melanoma patients who receive certain B-Raf inhibitors often develop secondary cancers driven by H-Ras mutations.”
Drugs that selectively target certain Ras-Raf binding pairs might offer improved therapeutic benefits, especially if the gene alterations detected in a cancer point to mutations in K-Ras, B-Raf or other Ras/Raf proteins, Morrison noted.
The researchers are now using BRET technology to screen for compounds that disrupt, either directly or indirectly, the Ras/Raf interaction in live cells. This approach could offer new strategies for developing more effective targeted therapies.