Learning how to design RNA-targeted therapeutics by manipulating a bacterial gene regulator.

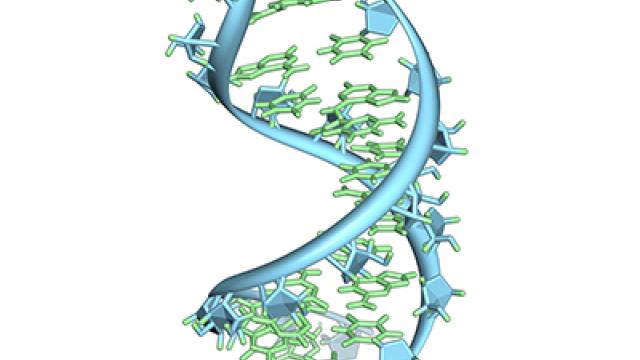
CCR scientists are developing small molecules that target RNA. This figure highlights a molecule interacting with a bacterial riboswitch that controls gene expression. Studying systems like this can provide the basis for developing new RNA-binding drugs.
Credit: Joseph Meyer, Scientific Publications, Graphics and Media, Frederick National Laboratory, NCI, NIH
One of life’s most essential and versatile molecules, RNA, is best known as DNA’s molecular messenger, relaying information encoded in the genome to the cellular machinery that builds proteins. However, only a small fraction of the RNA inside human cells plays that role. A vast assortment of additional RNAs perform a myriad of tasks that ultimately keep a wide range of cellular operations on track.
These activities exert a profound influence on health, and there is good evidence that dysregulation of RNA contributes to cancer and other diseases. Researchers expect that identifying ways to manipulate RNA molecules that contribute to disease could lead to a new generation of therapeutics.
John Schneekloth, Ph.D., is working to understand how small molecules interact with RNA. Collaborating with Adrian Ferré-D’Amaré, Ph.D., and colleagues at the National Heart, Lung, and Blood Institute (NHLBI), his team has engineered a molecule that potently activates a specific gene-regulating RNA found in bacteria. Their success, reported in Cell Chemical Biology, offers important clues into the discovery and design of molecules that interact with RNA targets and could point the way toward development of a novel class of antibiotics.
The synthetic molecule developed by the team targets a gene-regulating segment of RNA called a riboswitch. Riboswitches, which are found primarily in bacteria, act as cellular sensors that can be activated by small molecules, or ligands. When a riboswitch encounters and binds to its ligand, it adopts a new shape that changes the activity of its associated gene. Because they are more amenable to structural analysis than most other RNAs, riboswitches are useful models for understanding how RNA folds and interacts with other molecules, Schneekloth says.
Schneekloth and his colleagues set out to find a way to activate genes controlled by a group of riboswitches called ZTP riboswitches. They tested about 22,000 small molecules for their ability to bind to these switches and found some that did, but none that had the potent effects they were hoping for. So they tried a new strategy: if they couldn’t find a molecule that strongly activated the riboswitch, they would make one.
Rather than starting from scratch, the team turned to the riboswitch’s natural ligand, a metabolite called ZMP, and reengineered it. To guide their modifications of the natural molecule, NHLBI fellow Christopher Jones, Ph.D., generated a detailed structure of a ZMP-bound riboswitch. That structure revealed exactly how the metabolite nestles inside the folded RNA and suggested places for the team to tinker with ZMP to improve the interaction.
When the researchers administered their synthetic ligand to bacteria, it stimulated the activity of the ZTP riboswitch’s target genes more potently than the natural ligand. The achievement represents a new way to engineer molecules that can control gene expression in bacteria. More broadly, it also gives researchers a deeper understanding of how to target not just riboswitches, but other types of RNA. This could help guide the development of new therapeutics that interact with RNA molecules to treat cancer and other diseases.