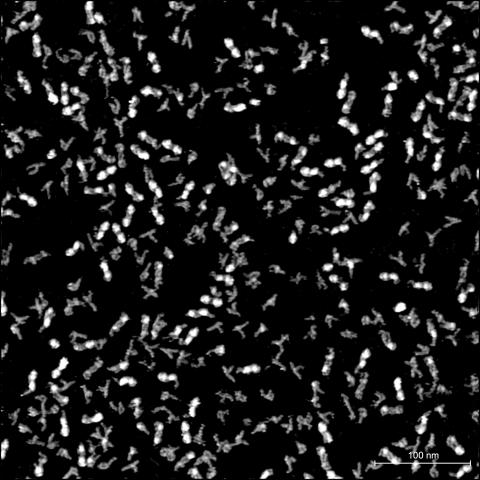
A high-resolution atomic force microscopy (AFM) image showing multiple conformations of identical RNA sequences. Image credit: Wang Lab
A single RNA sequence can fold into more than one biologically active shape, according to a recent study led by Yun-Xing Wang, Ph.D., Senior Investigator in the Center for Structural Biology. This finding, published February 9, 2023, in Nature Communications, challenges the current dogma that one sequence conforms to only one three-dimensional structure, leading to direct implications for RNA-targeted drug design in cancer and other diseases.
“This is a very significant advance in RNA biology,” says Wang, who has dedicated his career to understanding the relationship between RNA structure and function. For years, indirect measurements have suggested that RNA exists in dynamic conformations, but evidence of this phenomenon has not been directly observed until now. This is important, Wang explains, because one approach to drug design relies on understanding the architectural features of a drug’s target.
To look at single RNA molecules directly, postdoctoral fellow Jienyu Ding, Ph.D., used atomic force microscopy (AFM), a powerful tool that can visualize individual RNA molecules in liquid that is similar to the inside of a cell. This is in stark contrast to other methods for RNA structure determination — such as cryo-electron microscopy, crystallography or nuclear magnetic resonance — which average signals from thousands to quadrillions of RNA molecules observed under non-cell-like conditions.
Using AFM, the researchers examined vitamin B12 riboswitch RNA and found that identical RNA sequences can take on many conformations. They dubbed the conformations candy-shaped, y-shaped, p-shaped and compact, based on their familiar appearances. Importantly, the conformations behaved differently when given the opportunity to bind to vitamin B12, suggesting structure-dependent activity for a single RNA sequence.
More than 80% of the human genome is transcribed into RNA, which can control when, where and how efficiently genes are expressed in the cell, Wang emphasizes. “There are many RNA structures related to diseases,” he continues, “but researchers around the world have found that it’s challenging to design drugs against RNA due to a lack of RNA structures in databases. We don’t know what they look like.”
The team’s next step is to develop a complete and robust tool that researchers can use to determine the multiple 3D structures of a single RNA sequence. “Fundamentally, we want to change the belief that one sequence means one structure, and this could lead to a novel strategy for designing drugs against RNA targets,” Wang says.