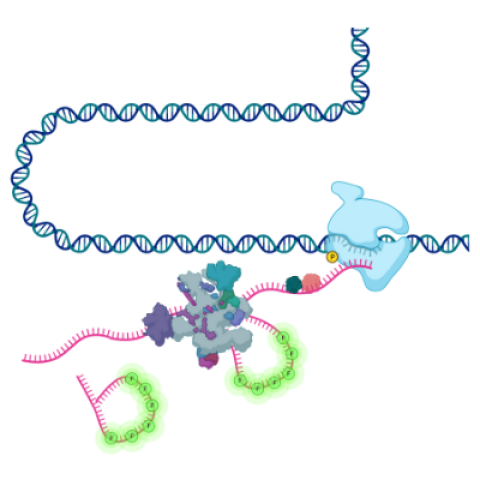
Schematic showing the transcription of DNA into RNA by RNA polymerase followed by splicing of the RNA by the spliceosome. Using single-molecule fluorescence imaging in living cells, Wan et al. was able to visualize the process of intron removal in real time.
Image credit: Yihan Wan, Research Fellow, LRBGE
CCR scientists have discovered that an essential step in gene expression — the removal of non-coding sequences from messenger RNA molecules — is more variable than previously thought. Researchers led by Daniel R. Larson, Ph.D., Senior Investigator in the Laboratory of Receptor Biology and Gene Expression, used live-cell imaging to visualize sequences known as introns being removed from individual molecules of messenger RNA. Their findings, reported May 11, 2021, in Cell, offer a new view of how cells process RNA to prepare for protein synthesis.
Introns are embedded within the genetic instructions for building proteins in all but the simplest organisms. These interrupting sequences are retained when a gene is transcribed into an RNA copy, or transcript, but must be removed before the transcript can be used to produce functional proteins.
Most human genes include around eight introns, but some genes contain over 100. A large, dynamic enzyme, called the spliceosome, is responsible for finding and removing introns from every RNA transcript. Each time an intron is spliced, spliceosome components come together to assemble directly on the RNA molecule. Constant assembly and disassembly make it difficult to study some aspects of the splicing process, but new high-resolution methods for imaging living cells made it possible for Larson’s team to watch individual RNA molecules be synthesized and processed in real time.
Yihan Wan, Ph.D., a research fellow in Larson’s lab, developed a method that allowed her to observe bursts of transcription from hundreds of individual genes in human cells, as well as to monitor individual, fluorescently labeled introns over many hours. After following the processing of RNA transcripts from hundreds of human genes, she found there was a great deal of variability in the time it took the spliceosome to remove any particular intron. In different copies of the same transcript, the same intron might take minutes or hours to be removed. The team concluded that introns are not removed in a predetermined order; rather, the selection of splice sites by the spliceosome exhibits tremendous randomness.
What’s more, Wan says, they found far more splicing was taking place than researchers had presumed. Their experiments showed that rather than excising intact introns by cutting at each end, spliceosomes often make cuts inside introns, removing them in a multistep process. Larson says the finding that spliceosomes act at locations not previously considered by scientists to be splice sites substantially changes the understanding of how these enzymes do their work.