Studies of the pediatric cancer rhabdomyosarcoma suggest a way to exploit the complexity of gene networks to intervene in cancer growth.
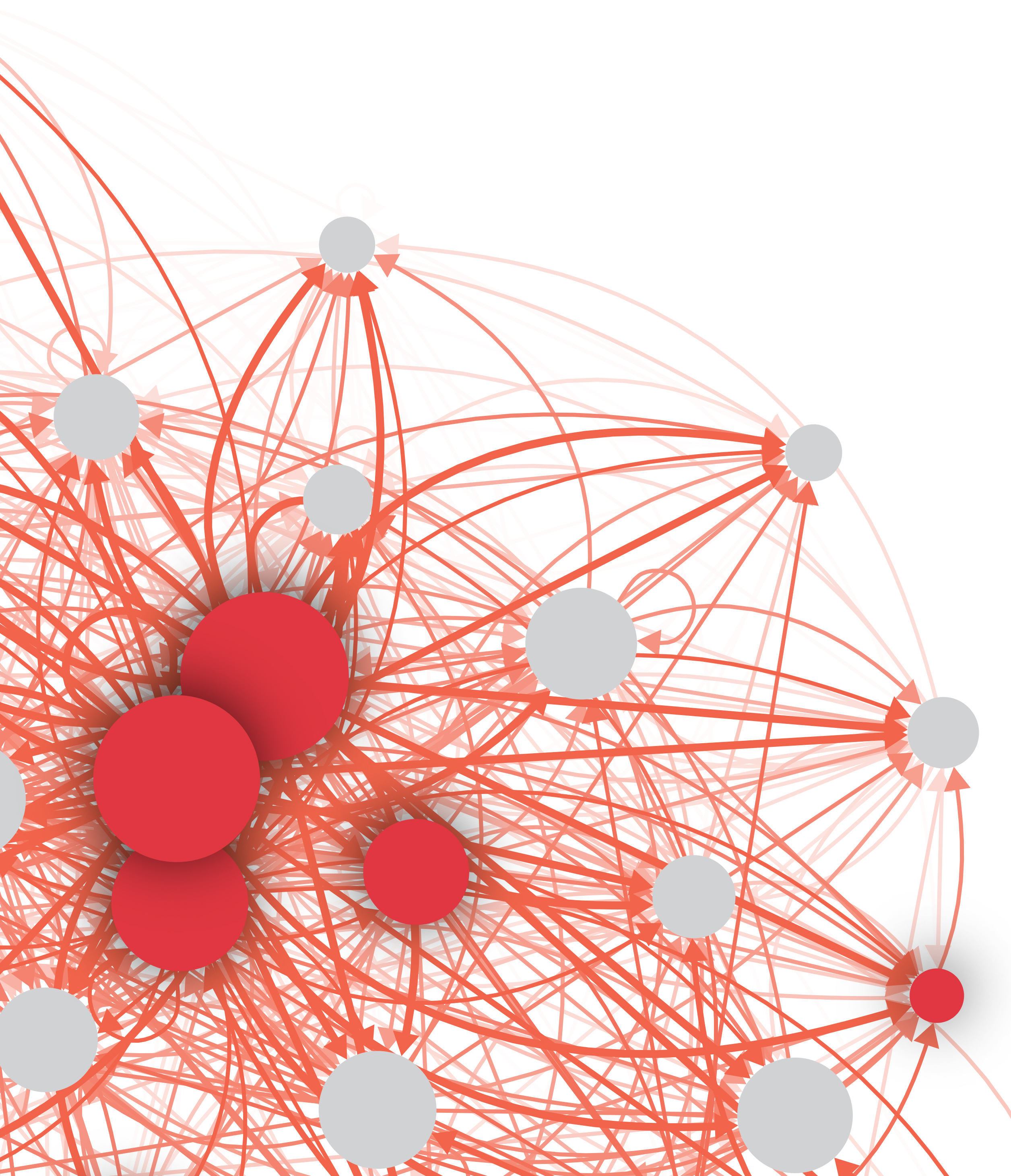
This network plot shows how each transcription factor (represented by a red or gray circular node) is connected to other transcription factors, forming groups that regulate genes that define PAX3-FOXO1 fusion-positive rhabdomyosarcoma. Each connection (represented by an arrow) is made by identifying a transcription factor’s DNA binding sequence in the super enhancer that controls another transcription factor.
Credit: Berkley Gryder, CCR, NCI, NIH
The growth of most cancer cells is fueled by networks of genes, yet therapeutic strategies often target only a single gene. New research from CCR scientists suggests that people with rhabdomyosarcoma may benefit from a class of drugs known as histone deacetylase inhibitors, which slows the activity of an entire set of cancer-driving genes.
Rhabdomyosarcoma, a cancer most often diagnosed in children, develops in the body’s soft tissues and evolves most commonly from immature muscle cells. Aggressive cases of the disease are often driven by a rearrangement within the genome known as a PAX fusion, which occurs when parts of two chromosomes fuse together. This change disrupts a gene network that guides the development of fully functional muscle cells. When a PAX fusion is present, muscle-cell progenitors fail to mature properly and grow excessively instead.
Javed Khan, M.D., realized that the PAX fusion presented an exploitable vulnerability. The disrupted genes that drive the growth of these cancers are part of a core-regulatory circuit, a type of network whose genes are regulated differently than other genes. Because of this, Khan suspected there might be a way to selectively shut down the PAX-fusion-regulated circuit in rhabdomyosarcoma cells. That, he hoped, might halt their growth.
Working with CCR’s Molecular Targets Laboratory, Khan’s team screened tens of thousands of compounds, seeking any that disabled the core-regulatory circuitry of rhabdomyosarcoma cells without disturbing other genes. According to results published in Nature Communications, one of the drug classes that was highly potent included small molecules referred to as histone deacetylase (HDAC) inhibitors. These compounds alter cells’ ability to process and interpret acetylation, a chemical modification that occurs on the histone proteins that organize DNA.
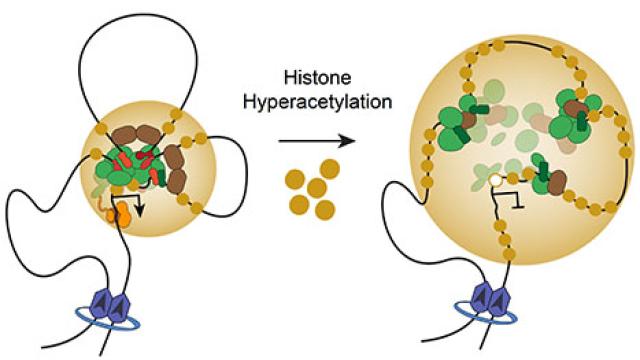
Inhibitors of histone deacetylation are known to affect how the genome is folded inside cells. In intricate work, Khan and his team discovered that HDAC inhibitors interfered with a mode of gene regulation that requires DNA to form a loop, bringing a distant regulatory region close to its target genes. By mapping the three-dimensional shape of the genome, they showed that HDAC inhibitors prevented DNA from assuming the correct organization, thereby preventing activation of PAX-fusion-controlled genes. Importantly, this effect was not limited to a single gene but affected an entire network of genes that are misregulated in rhabdomyosarcoma. The team reported these findings in Nature Genetics.
Khan says this new understanding gives researchers the information they need to begin exploring whether histone deacetylase inhibitors, likely in combination with other drugs, can treat rhabdomyosarcoma in patients who do not respond well to current therapies. He is also optimistic that this strategy for targeting entire networks of genes may be an effective way to treat other types of transcription-driven cancers.