Thomas Gonatopoulos-Pournatzis, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 560, Room 21-102B
- Frederick, MD, 21702-1201
- 301-846-6095
- 301-846-6911
- thomas.gonatopoulos@nih.gov
RESEARCH SUMMARY
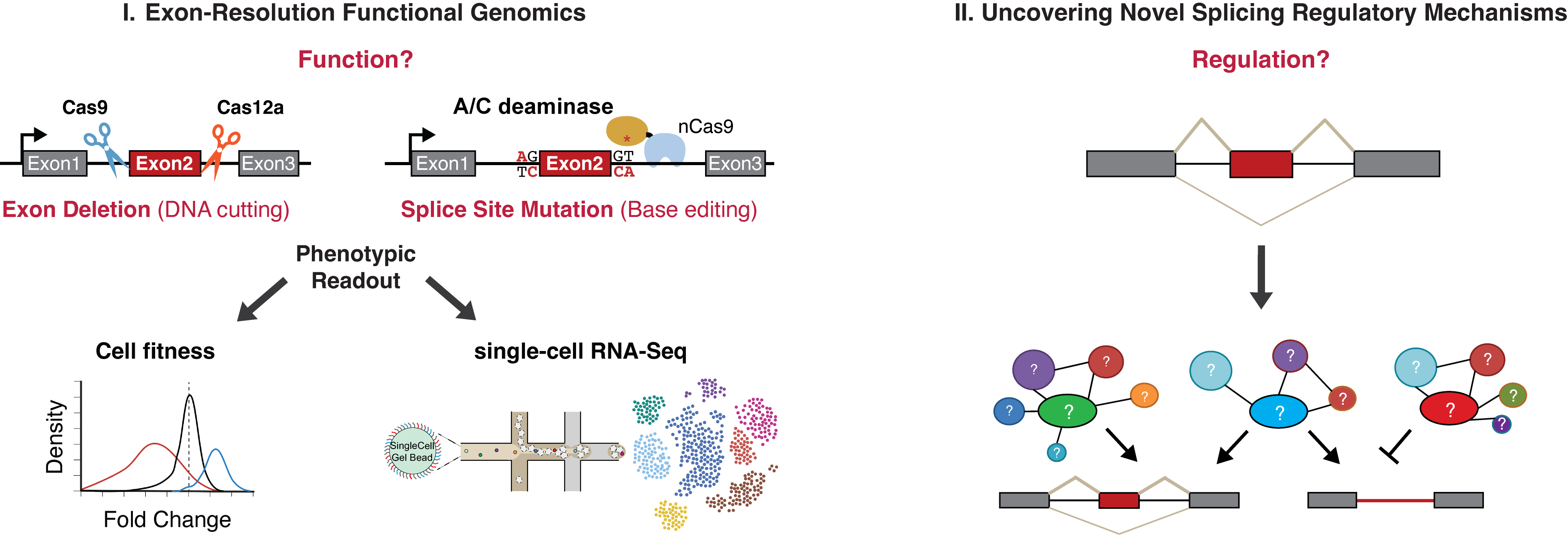
Thomas Gonatopoulos-Pournatzis is a leading molecular biologist who has pioneered functional genomics strategies to study the function and regulation of alternative splicing. His laboratory has developed innovative CRISPR-based screening approaches to dissect critical alternative exons, introns, and other RNA processing events. By integrating CRISPR genetic screens with diverse phenotypic analyses, including cell fitness, splicing reporters, and single-cell transcriptomics, his team systematically explores the regulatory and functional complexity of pre-mRNA processing. The ultimate aim of his research is to harness these insights to devise novel therapeutic strategies for cancer and other diseases.
Areas of Expertise

Thomas Gonatopoulos-Pournatzis, Ph.D.
Research
Aim 1: Identify and Characterize Phenotypically Relevant Alternatively Spliced Exons
A primary objective of our research is to systematically identify and characterize alternative exons with significant phenotypic roles. Aberrant splicing is recognized as a hallmark of cancer. By developing and leveraging innovative functional genomics platforms, including orthogonal exon deletion and splice site mutation strategies, we dissect the functional relevance of alternative exons at a genome-wide scale. We have identified numerous alternative exons that influence cell fitness and proliferation. Our team aims to highlight specific exons whose deletion creates vulnerabilities in cancer cells. Additionally, we are advancing exon deletion screening technology, aiming to integrate these platforms with high-content phenotypic readouts enabled by recent advances in single-cell analytics.
Aim 2: Map Novel Regulatory Networks that Control Alternative Splicing
While substantial strides have been made in deciphering the fundamental mechanisms of splicing, the intricate regulatory factors and pathways governing many alternative splicing choices remain inadequately characterized, especially in disease-state. Our approach integrates functional genomics tools, transcriptomics, computational analyses, biochemical methodologies, and animal models to delineate novel splicing regulatory factors and mechanisms and elucidate their in vivo significance and functional implications.
Relevance to cancer
Cancer cells exhibit widespread alterations in alternative splicing patterns. While substantial progress has been made in documenting these changes, identifying the downstream events responsible for disease development and progression often remains contentious. Our developed exon-resolution functional genomics strategies offer promising solutions to address this significant challenge in biomedical research.
Publications
Biography

Thomas Gonatopoulos-Pournatzis, Ph.D.
Dr. Gonatopoulos-Pournatzis earned his Bachelor's degree in Biology from the National and Kapodistrian University of Athens, Greece, in 2007, followed by a Master's degree from King's College London in 2008. He completed his Ph.D. in 2013 at the University of Dundee, Scotland, where he conducted research in Prof. Victoria Cowling's laboratory, focusing on the mechanisms underlying mRNA cap methylation and gene expression. For his postdoctoral studies, Dr. Gonatopoulos-Pournatzis joined Prof. Ben Blencowe's laboratory at the University of Toronto, Canada, where he explored the role of alternative splicing in neurons. His work was supported by prestigious fellowships from the European Molecular Biology Organization (EMBO), Canadian Institutes of Health Research (CIHR), and the Ontario Institute of Regenerative Medicine (OIRM). In 2020, Dr. Gonatopoulos-Pournatzis established the Functional Transcriptomics Section at the RNA Biology Laboratory of the National Cancer Institute (NIH). His research group integrates functional genomics and RNA biology to systematically investigate the regulation and function of alternative splicing programs in health and disease. Dr. Gonatopoulos-Pournatzis has received several honors, including the Baxter Prize (2012), the Donnelly Centre Research Excellence Award (2018), and the NIH Distinguished Scholar Program Award (2020).
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Lab Life

Summer gathering of our and Aregger lab following scavenger hunt through Downtown Frederick (2024).
Lab family trip to Maryland's coast line, Ocean City (2024).

After lunch we went for bowling in our 2023 holiday gathering.

Mission accomplished in the escape room! Kids in Frederick will also get the gifts from Santa this year (2022).

We say farewell to Shreya and Tyler by doing a Zipline trip in Harper's Ferry (2023)

Dinner at the historic town of Harper's Ferry (2023)

Celebrating RNA day with delicious home-made cookies baked by Shreya! (2022)

Group picture at the annual RNA Biology picnic (2023)

Wrapping up 2022 with a group lunch in Downtown Frederick. Thanks for the hard work everyone.

Tree Trekking in Frederick (2021)

Farewell party for Ethan who's leaving us to start a PhD at UCSB. Good luck with your grad studies Ethan! (2022)

Visit of a pumpkin farm in Frederick County (2020)

Joint lunch with the Wu lab at the NCI-Frederick campus (2021)

Hanging out at the pond of NCI-Frederick campus with Shreya and Britnie after the joint lunch with the Wu lab (2021).
Kun's Farewell before starting her MD/PhD at Ohio State University (2021)

50 ft above ground - perfect spot for a nap... (2021)

My name is Xiao, Meisheng Xiao ;-) (2021)
Team
News
July 2025 - A proud moment: Amit, Arun, and JJ each won an NIH FARE travel award this year—three awards for our lab!
May 2025 - Our high-throughput exon deletion screen is highlighted in the 2025 CCR Milestones, which showcases some of the past year’s major scientific breakthroughs from the Center for Cancer Research.
November 2024 - Congratulations to Arun for winning a poster prize at the 2024 NCI RNA Biology Initiative Retreat!
July 2024 - We are thankful to Molecular Cell editors for featuring our recent manuscript with a “meet the authors” article. You can read more about our journey in science here.
June 2024 - We are thrilled to announce that our group's first research manuscript has been published in Molecular Cell. Special thanks to everyone involved!
June 2024 - We are thrilled to welcome Sandra Le to our team for the next year as part of the prestigious Werner H. Kirsten (WHK) High School Senior Student Internship Program!
June 2024 - We are excited to welcome Benista Owusu-Amo to our group for a NIH Summer Internship!
January 2024 - Our team is growing. We are excited to welcome Jeongjin (JJ) Kim as a new PostDoc to the group!
July 2023 - Congratulations to Arun for winning an NIH FARE travel award!
June 2023 - We welcome Nikhil Parab to our group for a NIH Summer Internship!
May 2023 - Congratulations to Shreya for winning an outstanding poster award at the 2023 Frederick Spring Research Festival!
December 2022 - Bandana Kumari has joined our team as a computational PostDoc. We're excited to welcome Bandana at NCI-Frederick!
October 2022 - We're happy to extend our group and welcome Amit Behera who is joining our group after having completed his Postdoc in Angela Brooks lab at UCSC.
August 2022 - Ethan starting his PhD studies at the University of California Santa Barbara. Well done Ethan! We will miss you.
June 2022 - We welcome Max Teszler to our group for a NIH Summer Internship!
May 2022 - Congratulations to Shreya for winning the first poster prize presentation at the 2022 Frederick Spring Research Festival!
September 2021 - A protocol paper describing the use of our combinatorial Cas9-Cas12a screening platform for genetic interaction mapping and exon resolution functional genomics is now published in Nature Protocols.
September 2021 - A computational method for analyzing combinatorial CRISPR screens spearheaded by Henry Ward in Chad Myers' lab at the University of Minnesota Twin Cities has been published alongside our CHyMErA screening protocol in Nature Protocols.
August 2021 - A warm welcome to Shreya Kordale who joins our team as a PostBac student.
June 2021 - Michael has been selected as a recipient of an NIH Stadtman Investigator tenure-track position and he will open his lab at the Molecular Targets Program at NCI-Frederick in October 2021. Congratulations Michael!!
May 2021 - Congrats to Timothy who won an 'Outstanding Poster Award' at the Postbac Poster Day 2021!
April 2021 - Kun has been accepted for an MD/PhD position at Ohio State University. Well done Kun!
March 2021 - We're excited to further extend our team and to welcome Arun Prasath Damodaran who is starting his PostDoc in our group.
March 2021 - Our group has been awarded a Center of Cancer Research FLEX Technology Development Grant for developing a scalable combinatorial screening platform for high-content phenotyping. We’re very grateful for the immense support of NCI/NIH of our research!
November 2020 - We're excited to welcome Meisheng Xiao who is joining our group after having completed his Postdoc in Jeremy Wilusz lab at UPenn.
September 2020 - After several weeks of quarantine and waiting for biosafety approval we are now able to officially open the lab. Welcome to the team Michael, Ethan, Kun and Timothy and thanks everyone at NCI-Frederick and the RNA Biology Laboratory for their gracious welcoming!