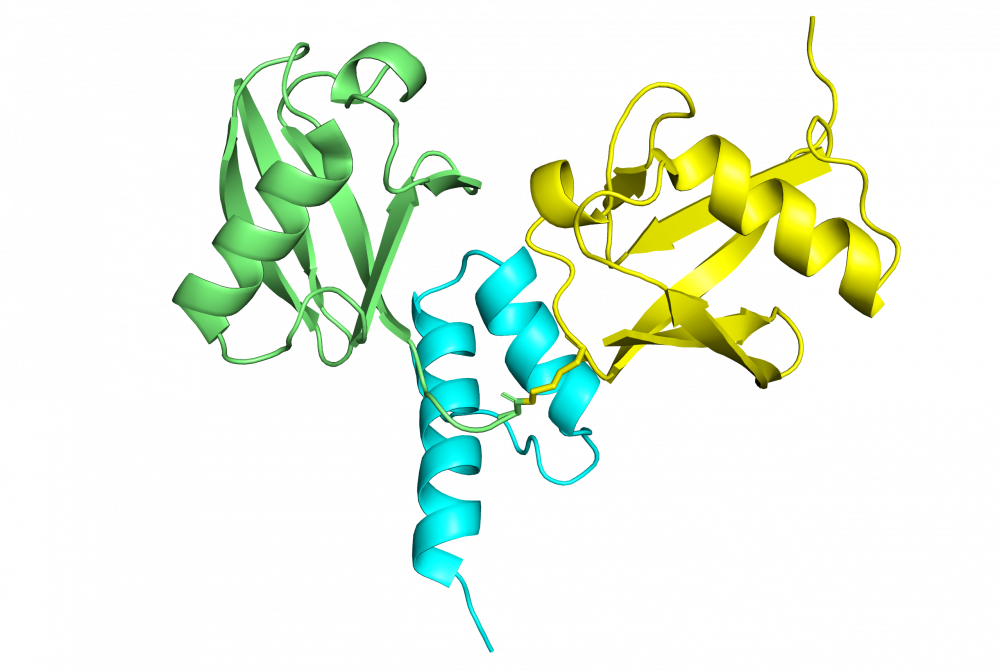Protein Processing Section
Kylie J. Walters, Ph.D.
Research
Dr. Walters' lab, the Protein Processing Section, studies the structural and mechanistic basis of ubiquitin signaling events, proteasome function, and protein quality control. Our lab uses an interdisciplinary approach that merges functional cell-based assays with a variety of biophysical and structural biology techniques to define the mechanistic underpinnings of how protein lifetimes are regulated in cells. We have helped to identify the mechanisms of substrate recognition by the proteasome, identifying Rpn1 and Rpn13 as substrate receptors that bind to ubiquitin chains directly and to the shuttle factors that deliver ubiquitinated substrates to the proteasome. By using NMR spectroscopy, we have solved structures of these receptors complexed with ubiquitin chains and shuttle factors. We have also used cryoelectron microscopy to study ubiquitin chain binding to the 26S proteasome.
Team
Covers

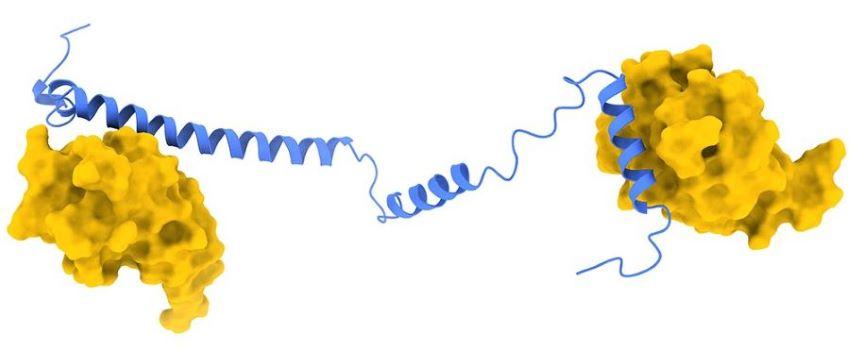
Pathogenic mechanisms of UBA1 patient mutations
Most cellular ubiquitin signaling is initiated by UBA1, which activates and transfers ubiquitin to tens of E2 enzymes. Clonally acquired UBA1 missense mutations cause an inflammatory-hematologic overlap disease called VEXAS (vacuoles, E1, X-linked, autoinflammatory, somatic) syndrome. Despite extensive clinical investigation into this lethal disease, little is known about the underlying molecular mechanisms. Here, by dissecting VEXAS-causing UBA1 mutations, we discovered that p.Met41 mutations alter cytoplasmic isoform expression, whereas other mutations reduce catalytic activity of nuclear and cytoplasmic isoforms by diverse mechanisms, including aberrant oxyester formation. Strikingly, non-p.Met41 mutations most prominently affect transthioesterification, revealing ubiquitin transfer to cytoplasmic E2 enzymes as a shared property of pathogenesis amongst different VEXAS syndrome genotypes. A similar E2 charging bottleneck exists in some lung cancer-associated UBA1 mutations, but not in spinal muscular atrophy-causing UBA1 mutations, which instead, render UBA1 thermolabile. Collectively, our results highlight the precision of conformational changes required for faithful ubiquitin transfer, define distinct and shared mechanisms of UBA1 inactivation in diverse diseases, and suggest that specific E1-E2 modules control different aspects of tissue differentiation and maintenance.

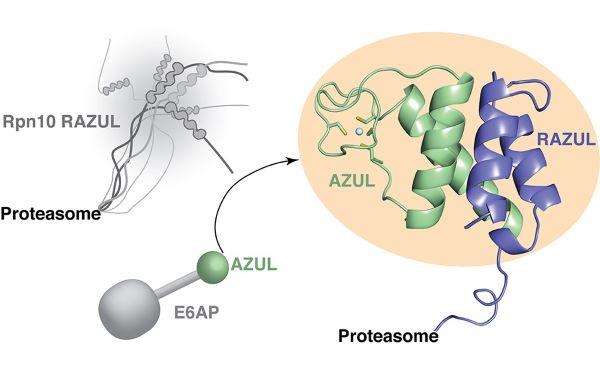
Proteasome-substrate interactions
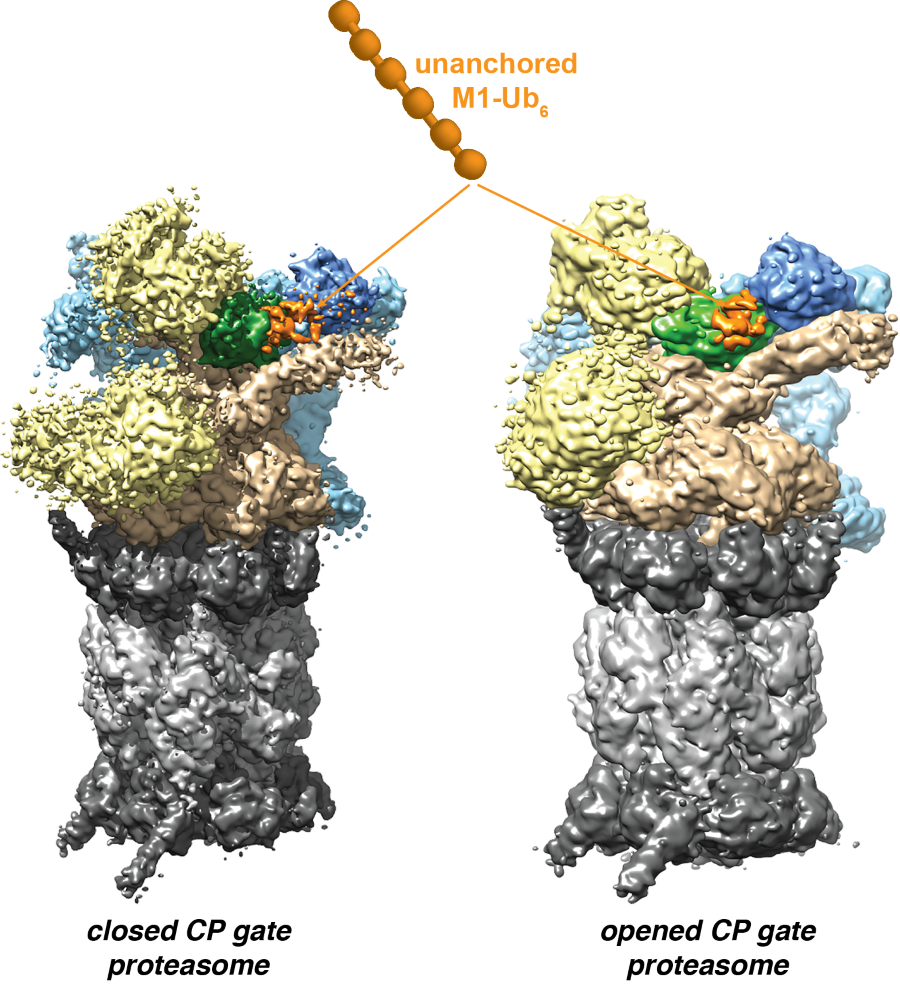
A top view of the EM density for the Rpt1-Rpt6 hexamer in the s4 state (EMDB: 9045), with the central channel of ATPase ring labeled. Image taken State-of-the-Art Review by Andreas Martin, Kylie Walters and co-authors on proteasome interactions with ubiquitinated substrates.

Impact of Losing hRpn13 Pru or UCHL5 on Proteasome Clearance of Ubiquitinated Proteins and RA190 Cytotoxicity
Region of the 26S proteasome (rendered as a ribbon display) highlighting the ubiquitin-binding T1 site (blue) of Rpn1 (orange) and absence of Rpn13 due to deletion of its Pru domain. Truncated Rpn13 (trRpn13) retains binding to deubiquitinating enzyme UCHL5, but not to the proteasome, causing loss of RA190-induced accumulation of ubiquitinated proteins. A ubiquitinated substrate is modeled as a surface diagram. Joseph Meyer and Xiuxiu Lu are acknowledged for assistance with this image.
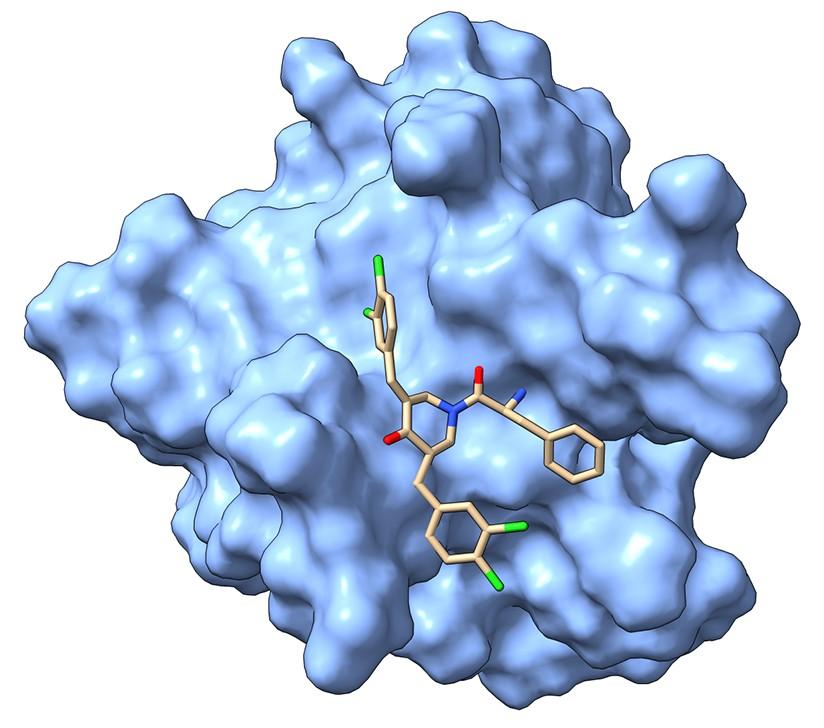
A bis-Benzylidine Piperidone Targeting Proteasome Ubiquitin Receptor RPN13/ADRM1 as a therapy for Cancer
The bis-benzylidine piperidone RA190 covalently binds to cysteine 88 of ubiquitin receptor RPN13 in the 19S regulatory particle and inhibits proteasome function, triggering rapid accumulation of polyubiquitinated proteins. Multiple myeloma (MM) lines, even those resistant to bortezomib, were sensitive to RA190 via endoplasmic reticulum stress-related apoptosis. RA190 stabilized targets of human papillomavirus (HPV) E6 oncoprotein, and preferentially killed HPV-transformed cells. After oral (p.o.) or intraperitoneal (i.p.) dosing of mice, RA190 distributed to plasma and major organs excepting brain, and inhibited proteasome function in skin and muscle. RA190 administration profoundly reduced growth of multiple myeloma and ovarian cancer xenografts, and oral RA190 treatment retarded HPV16+ syngeneic mouse tumor growth, without impacting spontaneous HPV-specific CD8+ T cell responses, suggesting its therapeutic potential.

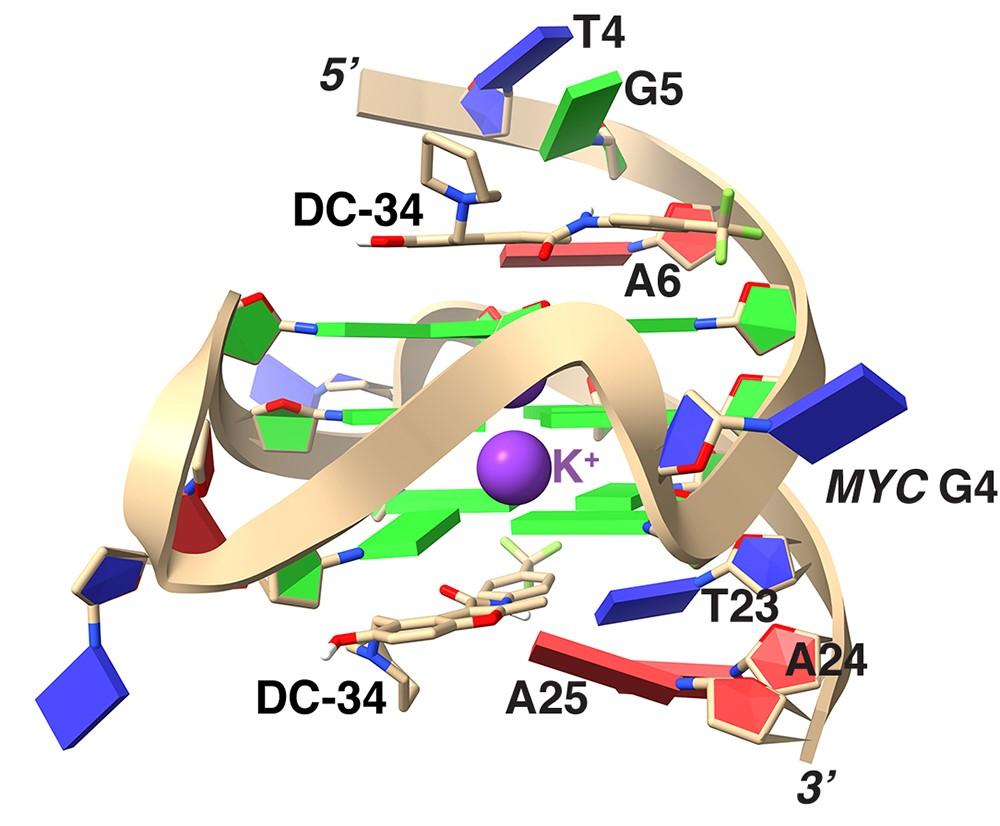
Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition
Ubiquitin is a key regulatory molecule in diverse cellular events. How cells determine the outcome of ubiquitylation remains unclear; however, a likely determinant is the specificity of ubiquitin receptor proteins for polyubiquitin chains of certain length and linkage. Proteasome subunit S5a contains two ubiquitin-interacting motifs (UIMs) through which it recruits ubiquitylated substrates to the proteasome for their degradation. Here, we report the structure of S5a (196-306) alone and complexed with two monoubiquitin molecules. This construct contains the two UIMs of S5a and we reveal their different ubiquitin-binding mechanisms and provide a rationale for their unique specificities for different ubiquitin-like domains. Furthermore, we provide direct evidence that S5a (196-306) binds either K63-linked or K48-linked polyubiquitin, and in both cases prefers longer chains. On the basis of these results we present a model for how S5a and other ubiquitin-binding proteins recognize polyubiquitin.
Alumni
Gallery
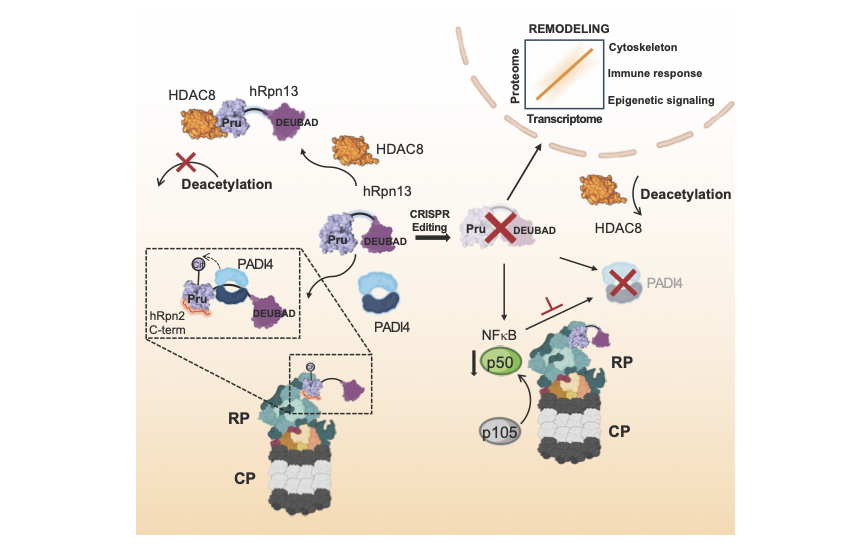
A representative schematic shows that PROTAC targeting or deletion of hRpn13 (purple) alters gene expression through hRpn13 interactions with epigenetic factors HDAC8 (orange), PADI4 (blue), and transcription factor NF-κB p50 (green) (Osei-Amponsa et al., Molecular Cell 84, 522-537, doi.org/10.1016/j.molcel.2023.11.035, (2024).
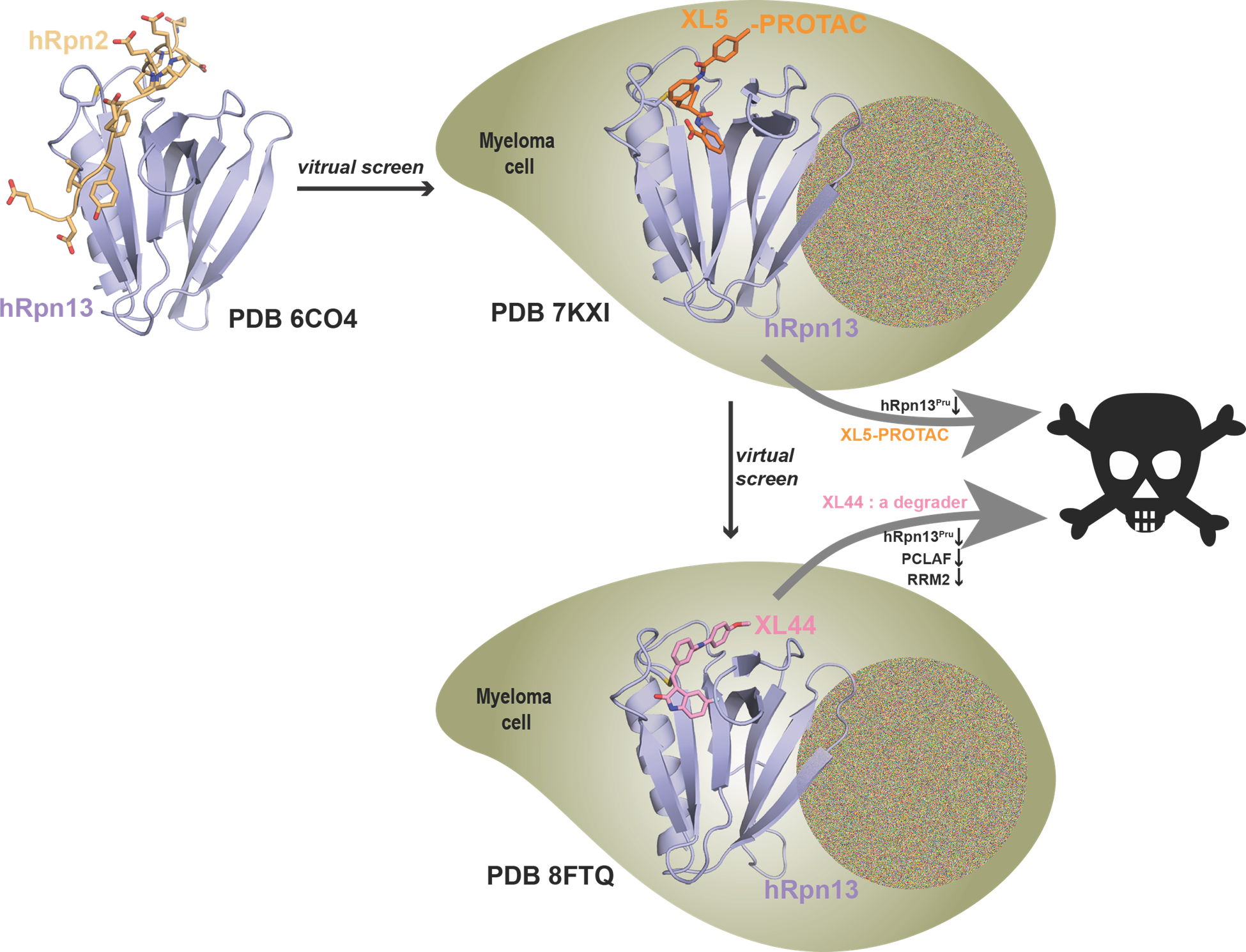
The NMR structure of hRpn13 Pru (periwinkle blue) with hRpn2 (940-953) (light orange) (PDB 6CO4; Lu, X. et al. Nat Commun. 2017 8, 15540) was used for a virtual screen to identify a new hRpn13-targeting scaffold with lead compound XL5. The NMR structure of XL5 (orange)-ligated hRpn13 Pru (periwinkle blue) (PDB 7KXI; Lu, X. et al. Nat Commun. 2021 12, 7318) was then used to design XL5-derived hRpn13 PROTACs including XL5-VHL-2. These PROTACs led to the discovery of an hRpn13 proteolytic product with an intact Pru domain (hRpn13Pru). hRpn13Pru is upregulated in myeloma patients and required for XL5-VHL-2 induced apoptosis (Nat Commun. 2021 12, 7318). To identify a more potent hRpn13-targeting compound, the hRpn13 Pru-XL5 structure was used in a covalent-docking screen to surprisingly identify the hRpn13 degrader XL44. The structure of XL44 (pink)-ligated hRpn13 Pru (periwinkle blue) was solved by integrated X-ray crystallography and NMR (PDB 8FTQ; Lu, X. et al. Nat Commun. 2024 15, 2485). In addition to degrading hRpn13Pru, XL44 also degrades a select few KEN-box proteins, including PCLAF and RRM2. However, XL44-induced apoptosis is hRpn13-dependent.
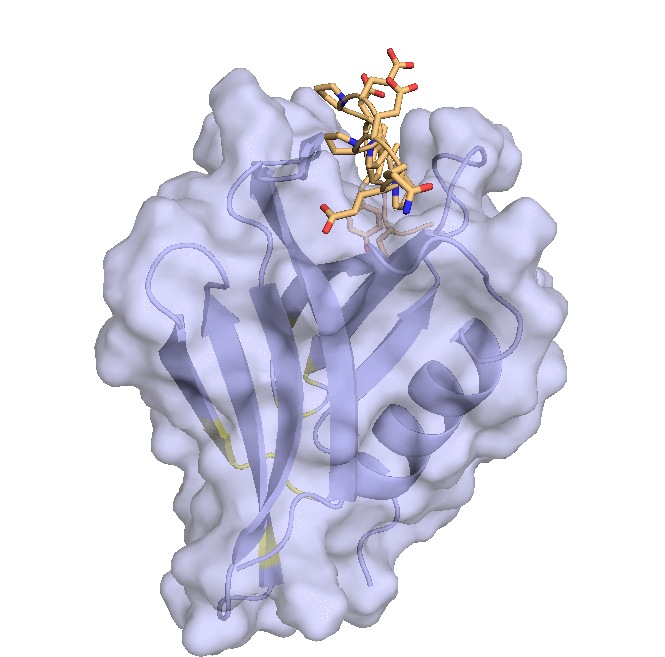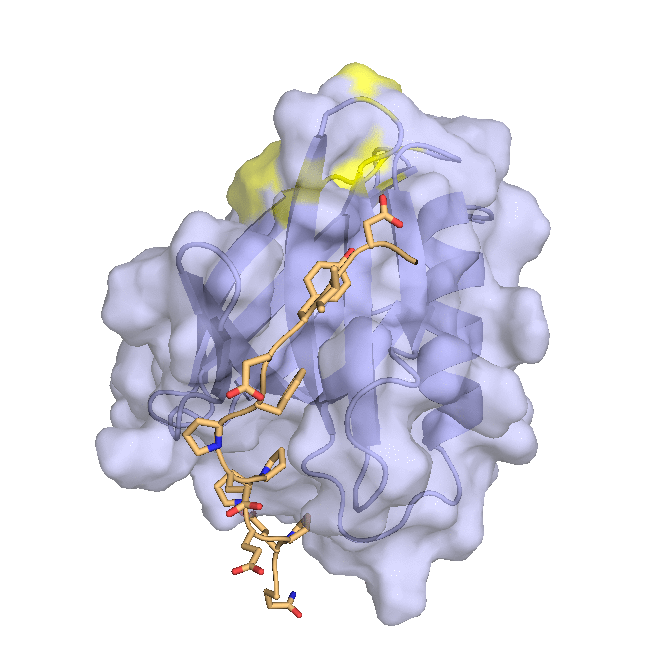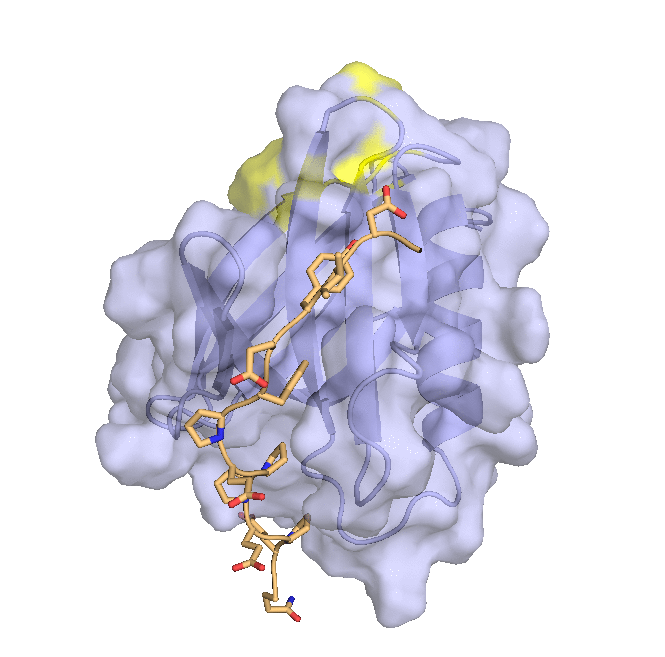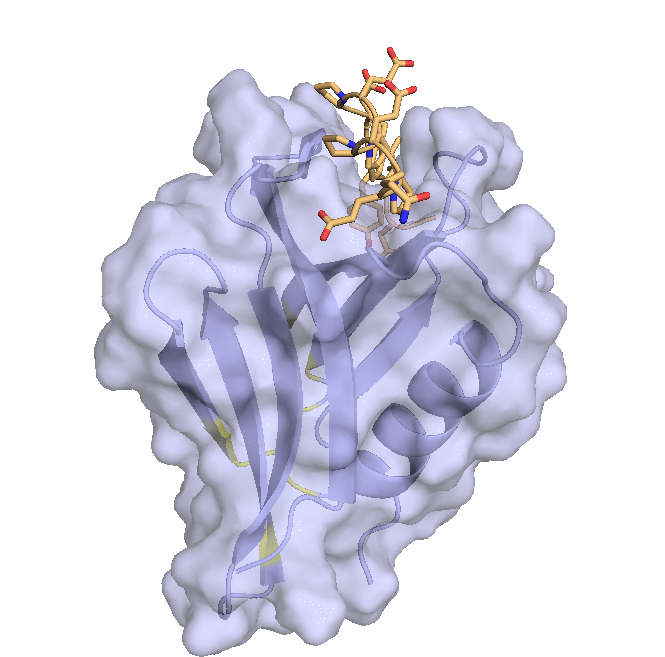
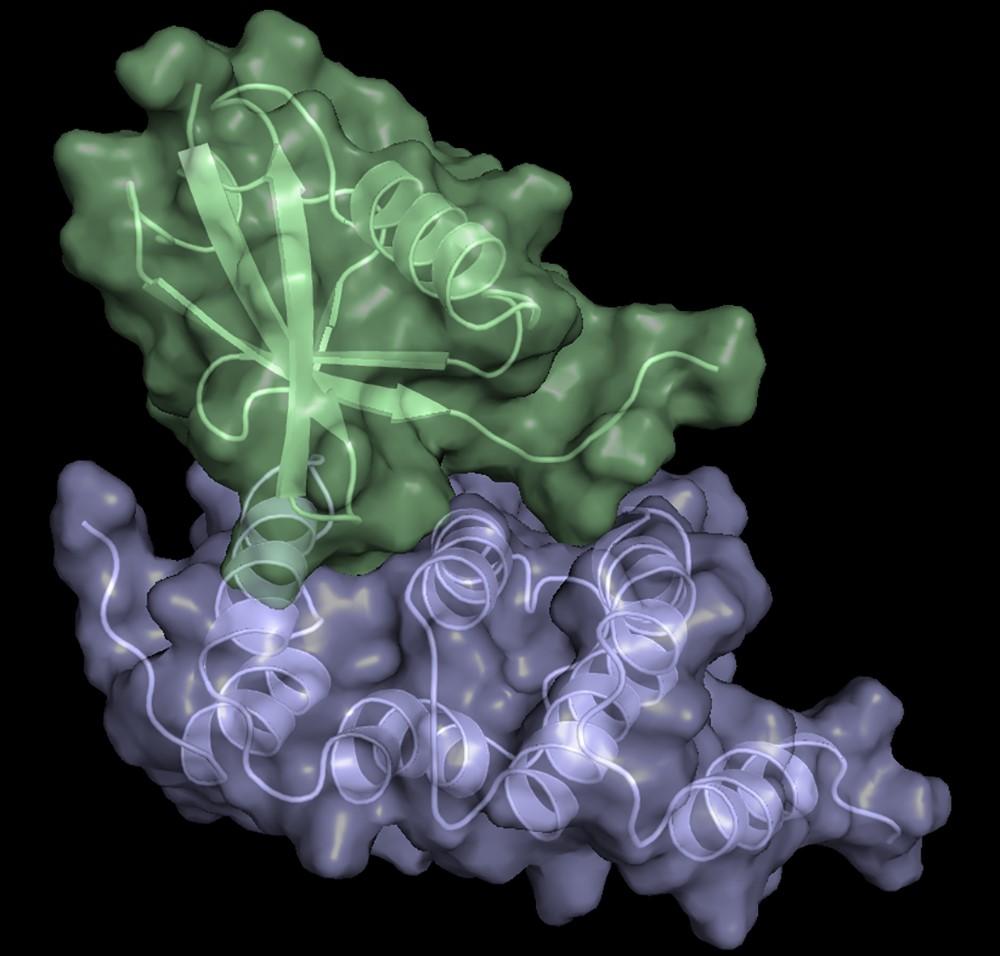
Structure of hRpn13 Pru with the 14-residue intrinsically disordered region at the extreme C-terminal end of hRpn2 that is used to recruit Rpn13 to the proteasome. The classic pleckstrin homology fold of hRpn13 Pru (periwinkle blue) is displayed as both a ribbon and surface diagram with hRpn2 (940-953) (light orange) as a stick model with nitrogen and oxygen atoms displayed in blue and red, respectively. The ubiquitin binding regions on hRpn13 Pru are colored yellow. (X. Lu et al. Nat Commun. 2017 8, 15540.)
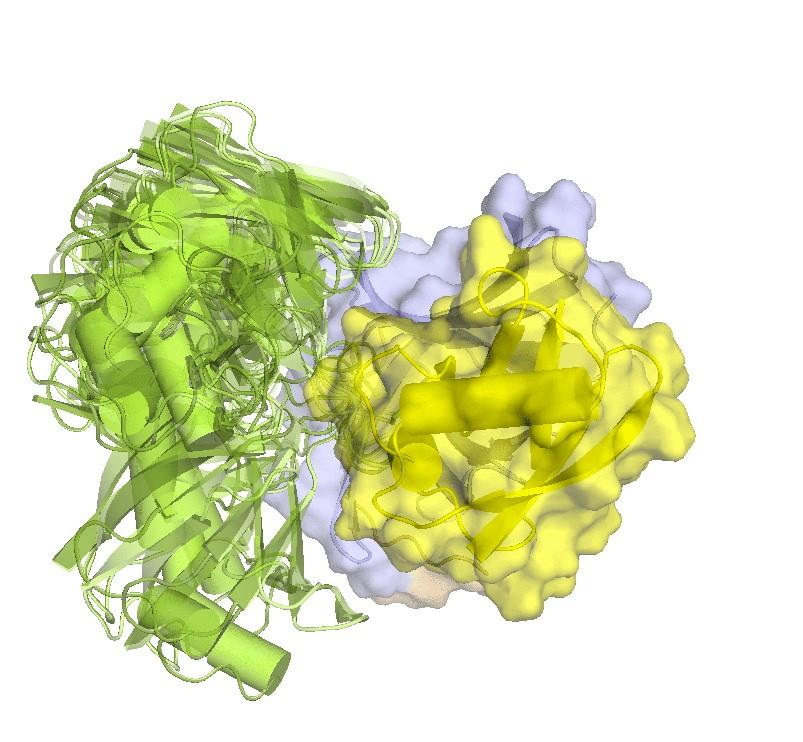
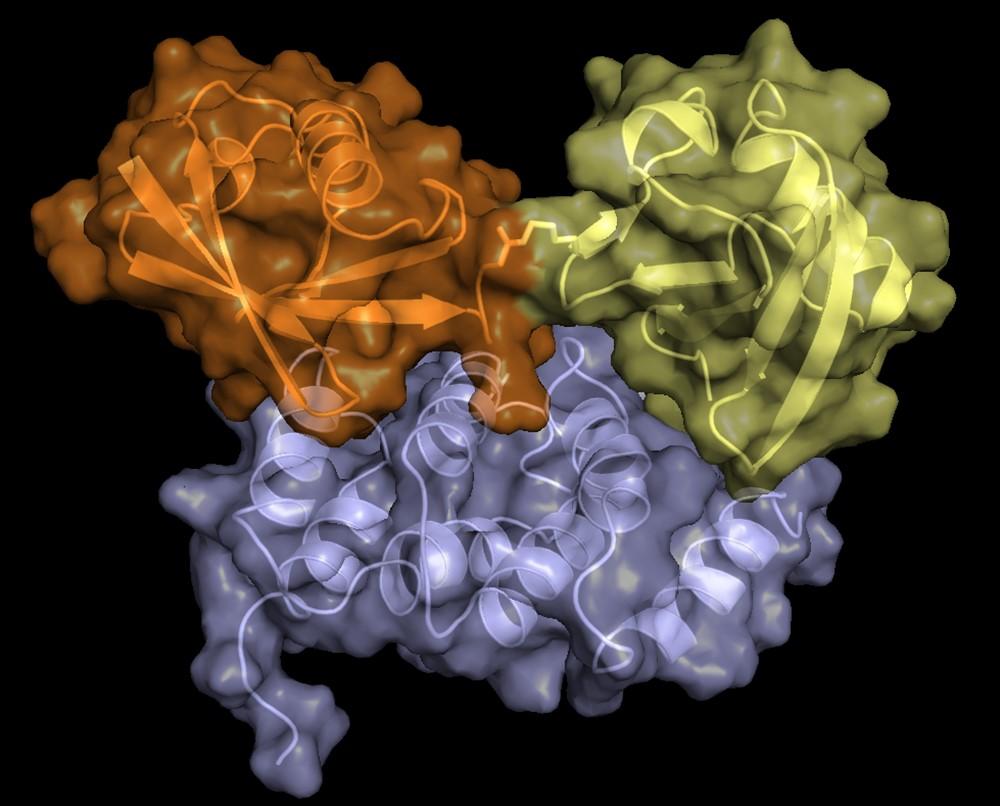
Structure of hRpn2-bound hRpn13 with K48-diubiquitin. hRpn13 Pru (periwinkle blue), hRpn2 (940-953) (light orange) and the bound proximal ubiquitin (yellow) of K48-diubiquitin are shown as a ribbon and surface diagrams. The full conformational ensemble is displayed for unbound distal ubiquitin (green) of K48-diubiquitin with each conformer as a transparent ribbon diagram (PDB: 6UYI) (X. Lu et al. Structure 28, 495-506 e493, doi:10.1016/j.str.2020.02.007 (2020).
Structure of hRpn13 Pru complexed with K48-diUb and the hRpn2 C-terminus.
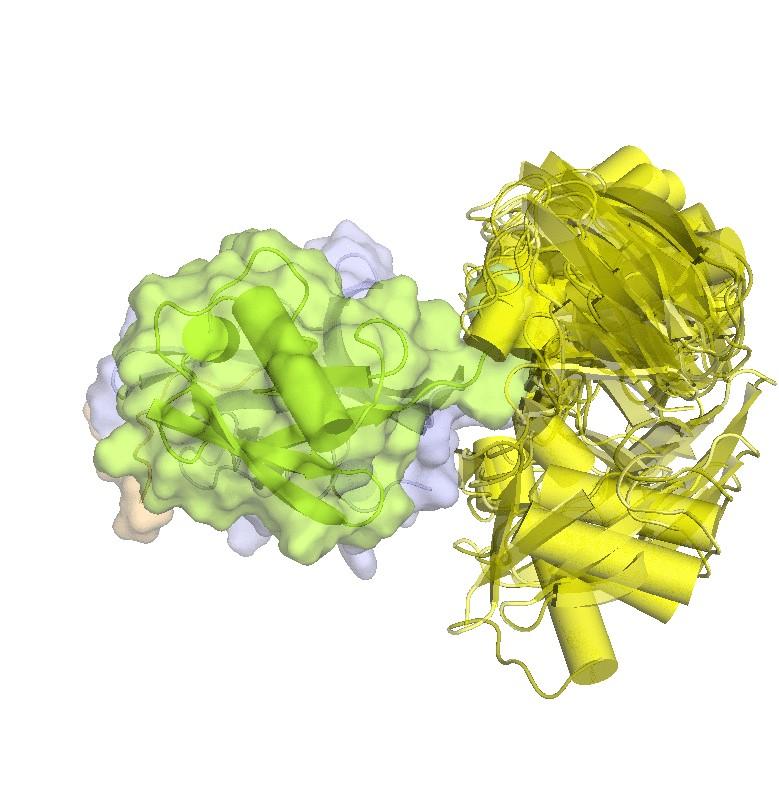
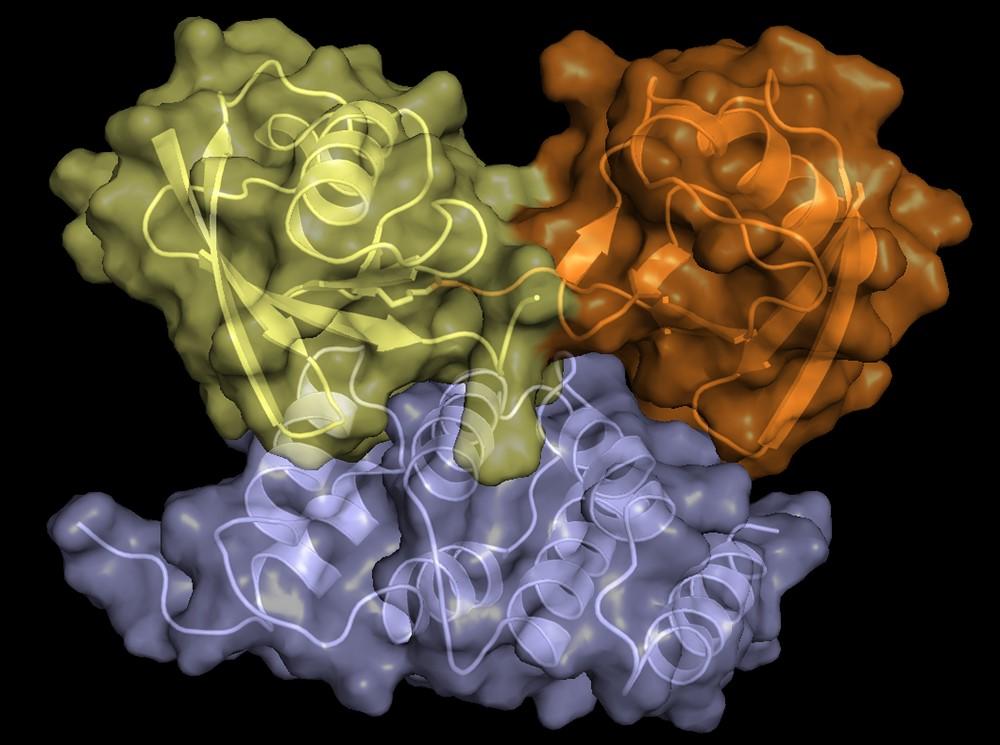
Structure of hRpn2-bound hRpn13 with K48-diubiquitin. hRpn13 Pru (periwinkle blue), hRpn2 (940-953) (light orange) and the bound distal ubiquitin (green) of K48-diubiquitin are shown as ribbon and surface diagrams. The full conformational ensemble of the unbound proximal ubiquitin (yellow) of K48-diubiquitin is displayed with each conformer as a transparent ribbon diagram (PDB: 6UYJ) (X. Lu et al. Structure 28, 495-506 e493, doi:10.1016/j.str.2020.02.007 (2020).
Structure of hRpn13 Pru complexed with K48-diUb and the hRpn2 C-terminus.