Proteasomes use a previously unknown method to recognize and process proteins that need to be destroyed.
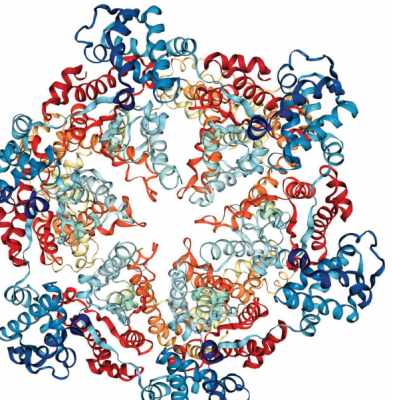
A proteasome is the cellular machine that disintegrates unwanted proteins. It’s a large, barrel-shaped complex with protein-degrading enzymes in its internal core. Credit: Protein Data Bank ID: 1DO2
For cells, disposing of proteins at the right time is as critical as producing essential proteins in the first place. Proteins that linger too long can be disruptive, as can those that are damaged or defective. Cells devote a lot of resources to identifying and destroying these unwanted proteins. The process is so essential that errors in the system have been linked to a range of diseases, including developmental disorders, neurodegenerative diseases, immune disorders and cancer.
The cellular machine that disintegrates unwanted proteins is called the proteasome, a large, barrel-shaped complex with protein-degrading enzymes in its internal core. A large fleet of enzymes patrols cells and marks proteins to be destroyed with a chemical tag that is recognized by the proteasome. This label ensures that proteasomes only destroy proteins that are faulty or unsuitable for a cell’s current conditions.
Kylie Walters, Ph.D., a Senior Investigator in CCR’s Structural Biophysics Laboratory, studies this regulated protein-disposal pathway. Her work has helped scientists develop a detailed understanding of how proteasomes recognize and degrade target proteins. This is important for cancer because proteasomes are thought to work overtime in tumor cells, which generate more unwanted proteins than normal cells. A cancer cell’s reliance on proteasomes to get rid of the excess proteins makes it more vulnerable to drugs that block this function. Since they were introduced in 2003, proteasome inhibitors, such as bortezomib (Velcade) and ixazomib (Ninlaro), have dramatically improved survival for patients with certain blood cancers. Researchers hope to extend this approach to treat other types of cancer.
In a study published in Science, Walters and her collaborators, Daniel Finley, Ph.D., from Harvard Medical School and John Engen, Ph.D., from Northeastern University, focused on how proteasomes recognize the chemical tag that marks proteins destined for degradation, a small molecule called ubiquitin. This recognition takes place at the cap of the proteasome, which processes target proteins and ensures only properly tagged molecules enter the core of the proteasome.
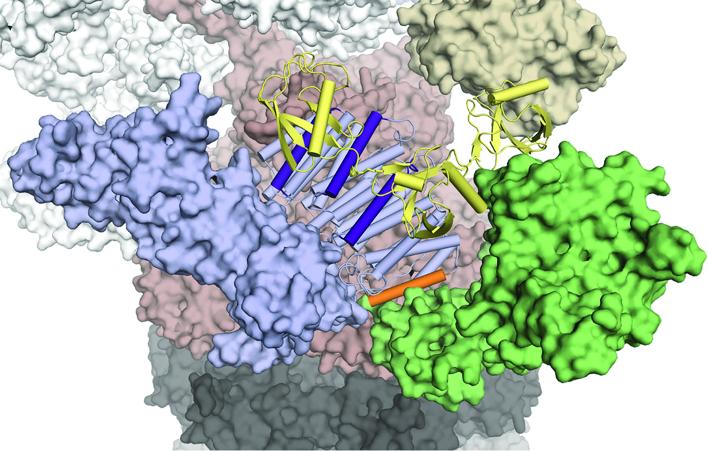
A newly identified ligand-binding hotspot in the proteasome for assembling substrates and cofactors. Credit: Xiang Chen, CCR, NCI, NIH
Walters and her colleagues discovered recognition sites for ubiquitin and structurally related molecules on a component of the proteasome cap called Rpn1. One of these sites can bind both to ubiquitin tags as well as a shuttle protein that escorts certain tagged proteins to proteasomes, while a second site binds to an enzyme that can remove ubiquitin tags. The tags must be trimmed away from a target protein before it can enter the interior of the proteasome, where it is degraded. According to the team’s findings, Rpn1 coordinates the proteasome’s recognition of target proteins with this deubiquitination step.
The work suggests a previously unknown way in which proteasomes recognize and process their protein targets. Understanding Rpn1’s role in this process should help researchers develop anticancer therapies that target the proteasome differently than existing inhibitors, which block enzymes in the interior of the proteasome. Walters says such drugs might be effective in patients whose cancers have become resistant to the existing drugs and might act synergistically when used in combination.
Shi Y, et al. Science. 2016 Feb 19;351(6275).