The primary focus of MTP is the collaborative creation, adaptation and use of state-of-the-art biomolecular screening technologies to discover compounds (including non-peptidic, peptidic and proteinaceous compounds) that selectively interact with or modify functions of specific molecular targets (e.g., endogenous molecules, structures, processes or pathways) of priority interest to the CCR, NCI. Tangible products of these efforts include novel or known compounds that may be used as "bioprobes" for target validation research or as drug discovery leads or templates.
Molecular Targets Program
Molecular Targets Program
About
The Molecular Targets Program (MTP) is a collaborative, team-science oriented laboratory engaged in discovery and translational sciences. The MTP provides the CCR a point of focus and infrastructure to enable CCR Investigators to pursue interdisciplinary, collaborative, applied, molecularly targeted drug discovery research within a matrix organizational format that is both supportive of, and complementary to, the traditional NCI/NIH intramural Lab/Branch organization. The MTP leverages the diverse, multidisciplinary expertise of its sections to facilitate the identification of genetic dependencies and potential therapeutic targets; the development of high-throughput screening assays to interrogate targets of interest; the discovery and optimization of bioactive compounds, including synthetic compounds, natural products, and peptides/proteins; and mechanistic studies on active compounds to support both basic and translational cancer research.
Job Vacancies
| Position | Degree Required | Contact Name | Contact Email |
|---|---|---|---|
| Postdoctoral Fellow - Drug Discovery, High Throughput Assays, Biosensor Development | Ph.D. or equivalent | Brice Wilson | MTP_PostDoc@nih.gov |
News
Learn more about CCR research advances, new discoveries and more
on our news section.
Covers

Acroamine A, a 2-Amino Adenine Alkaloid from the Marine Soft Coral Acrozoanthus australiae and Its Semisynthetic Derivatives That Inhibit cAMP-Dependent Protein Kinase A Catalytic Subunit Alpha
A high throughput screen performed to identify catalytic inhibitors of the oncogenic fusion form of cAMP-dependent protein kinase A catalytic subunit alpha (J-PKAcα) found an individual fraction from an organic extract of the marine soft coral Acrozoanthus australiae as active. Bioassay-guided isolation led to the identification of a 2-amino adenine alkaloid acroamine A (1), the first secondary metabolite discovered from this genus and previously reported as a synthetic product. As a naturally occurring protein kinase inhibitor, to unambiguously assign its chemical structure using modern spectroscopic and spectrometric techniques, five N-methylated derivatives acroamines A1–A5 (2–6) were semisynthesized. Three additional brominated congeners A6-A8 (7–9) were also semisynthesized to investigate the structure–activity relationship of the nine compounds as J-PKAcα inhibitors. Compounds 1–9 were tested for J-PKAcα and wild-type PKA inhibitory activities, which were observed exclusively in acroamine A (1) and its brominated analogs (7–9) achieving moderate potency (IC50 2–50 μM) while none of the N-methylated analogs exhibited kinase inhibition.

A new bispyrroloiminoquinone alkaloid from a Thai collection of Clavelina sp.
Two bispyrroloiminoquinone alkaloids, wakayin (1) and a new natural product 16-hydroxy-17-oxindole wakayin (2), were isolated from a Thai collection of the ascidian Clavelina sp. Herein, we present the isolation and structural elucidation of the new natural product and compare its NCI-60 cytotoxic activity to the known analogue wakayin and a series of other related pyrroloiminoquinones.
Grkovic T, Ruchirawat S, Kittakoop P, Grothaus PG, Evans JR, Britt JR, Newman DJ, Mahidol C, O'Keefe BR. A new bispyrroloiminoquinone alkaloid from a Thai collection of Clavelina sp. Asian J Org Chem 10: 1647-1649, 2021.

Agelasine diterpenoids and Cbl-b inhibitory ageliferins from the coralline demosponge Astrosclera willeyana
An extract of the coralline demosponge Astrosclera willeyana inhibited the ubiquitin ligase activity of the immunomodulatory protein Cbl-b. The bioassay-guided separation of the extract provided ten active compounds, including three new N-methyladenine-containing diterpenoids, agelasines W–Y (1–3), a new bromopyrrole alkaloid, N(1)-methylisoageliferin (4), and six known ageliferin derivatives (5–10). The structures of the new compounds were elucidated from their spectroscopic and spectrometric data, including IR, HRESIMS, and NMR, and by comparison with spectroscopic data in the literature. While all of the isolated compounds showed Cbl-b inhibitory activities, ageliferins (4–10) were the most potent metabolites, with IC50 values that ranged from 18 to 35 μM.
Jiang W, Wang D, Wilson BAP, Kang U, Bokesch HR, Smith EA, Wamiru A, Goncharova EI, Voeller D, Lipkowitz S, O'Keefe BR, Gustafson KR. Agelasine diterpenoids and Cbl-b inhibitory ageliferins from the coralline demosponge Astrosclera willeyana. Mar Drugs 19: 361, 2021.

Neopetrothiazide, an intriguing pentacyclic thiazide alkaloid from the sponge Neopetrosia sp.
Neopetrothiazide (1), a pentacyclic isoquinoline quinone, was isolated from a Neopetrosia sp. sponge. The structure elucidation was facilitated by utilizing long-range heteronuclear single quantum multiple bond correlation (LR-HSQMBC) and heteronuclear multiple bond correlation (HMBC) nuclear magnetic resonance (NMR) pulse sequences optimized to detect four- and five-bond 1H–13C heteronuclear correlations. These NMR experiments can help assign proton-deficient structural motifs like neopetrothiazide (1), which has 14 contiguous nonprotonated centers (C, N, and S). Neopetrothiazide (1), with an unprecedented thiazide-fused structural scaffold, is the first natural product containing a thiazide moiety.
Wang D, Jiang W, Kim CK, Bokesch HR, Woldemichael GM, Gryder BE, Shern JF, Khan J, O'Keefe BR, Beutler JA, Gustafson KR. Neopetrothiazide, an intriguing pentacyclic thiazide alkaloid from the sponge Neopetrosia sp. Org Lett 23: 3278-3281, 2021.

Molecular genomic features associated with in vitro response of the NCI-60 cancer cell line panel to natural products
Natural products remain a significant source of anticancer chemotherapeutics. The search for targeted drugs for cancer treatment includes consideration of natural products, which may provide new opportunities for antitumor cytotoxicity as single agents or in combination therapy. We examined the association of molecular genomic features in the well-characterized NCI-60 cancer cell line panel with in vitro response to treatment with 1302 small molecules which included natural products, semisynthetic natural product derivatives, and synthetic compounds based on a natural product pharmacophore from the Developmental Therapeutics Program of the US National Cancer Institute's database. These compounds were obtained from a variety of plant, marine, and microbial species. Molecular information utilized for the analysis included expression measures for 23059 annotated transcripts, lncRNAs, and miRNAs, and data on protein-changing single nucleotide variants in 211 cancer-related genes. We found associations of expression of multiple genes including SLFN11, CYP2J2, EPHX1, GPC1, ELF3, and MGMT involved in DNA damage repair, NOTCH family members, ABC and SLC transporters, and both mutations in tyrosine kinases and BRAF V600E with NCI-60 responses to specific categories of natural products. Hierarchical clustering identified groups of natural products, which correlated with a specific mechanism of action. Specifically, several natural product clusters were associated with SLFN11 gene expression, suggesting that potential action of these compounds may involve DNA damage. The associations between gene expression or genome alterations of functionally relevant genes with the response of cancer cells to natural products provide new information about potential mechanisms of action of these identified clusters of compounds with potentially similar biological effects. This information will assist in future drug discovery and in design of new targeted cancer chemotherapy agents.
Krushkal J, Negi S, Yee LM, Evans JR, Grkovic T, Palmisano A, Fang J, Sankaran H, McShane LM, Zhao Y, O'Keefe BR. Molecular genomic features associated with in vitro response of the NCI-60 cancer cell line panel to natural products. Molecular Oncology 15: 381-406, 2021.

Creating and screening natural product libraries
The National Cancer Institute of the United States (NCI) has initiated a Cancer Moonshot program entitled the NCI Program for Natural Product Discovery. As part of this effort, the NCI is producing a library of 1 000 000 partially purified natural product fractions which are being plated into 384-well plates and provided to the research community free of charge. As the first 326 000 of these fractions have now been made available, this review seeks to describe the general methods used to collect organisms, extract those organisms, and create a prefractionated library. Importantly, this review also details both cell-based and cell-free bioassay methods and the adaptations necessary to those methods to productively screen natural product libraries. Finally, this review briefly describes post-screen dereplication and compound purification and scale up procedures which can efficiently identify active compounds and produce sufficient quantities of natural products for further pre-clinical development.
Wilson BAP, Thornburg CC, Henrich CJ, Grkovic T, O'Keefe BR. Creating and screening natural product libraries. Nat Prod Rep 37: 893-918, 2020.
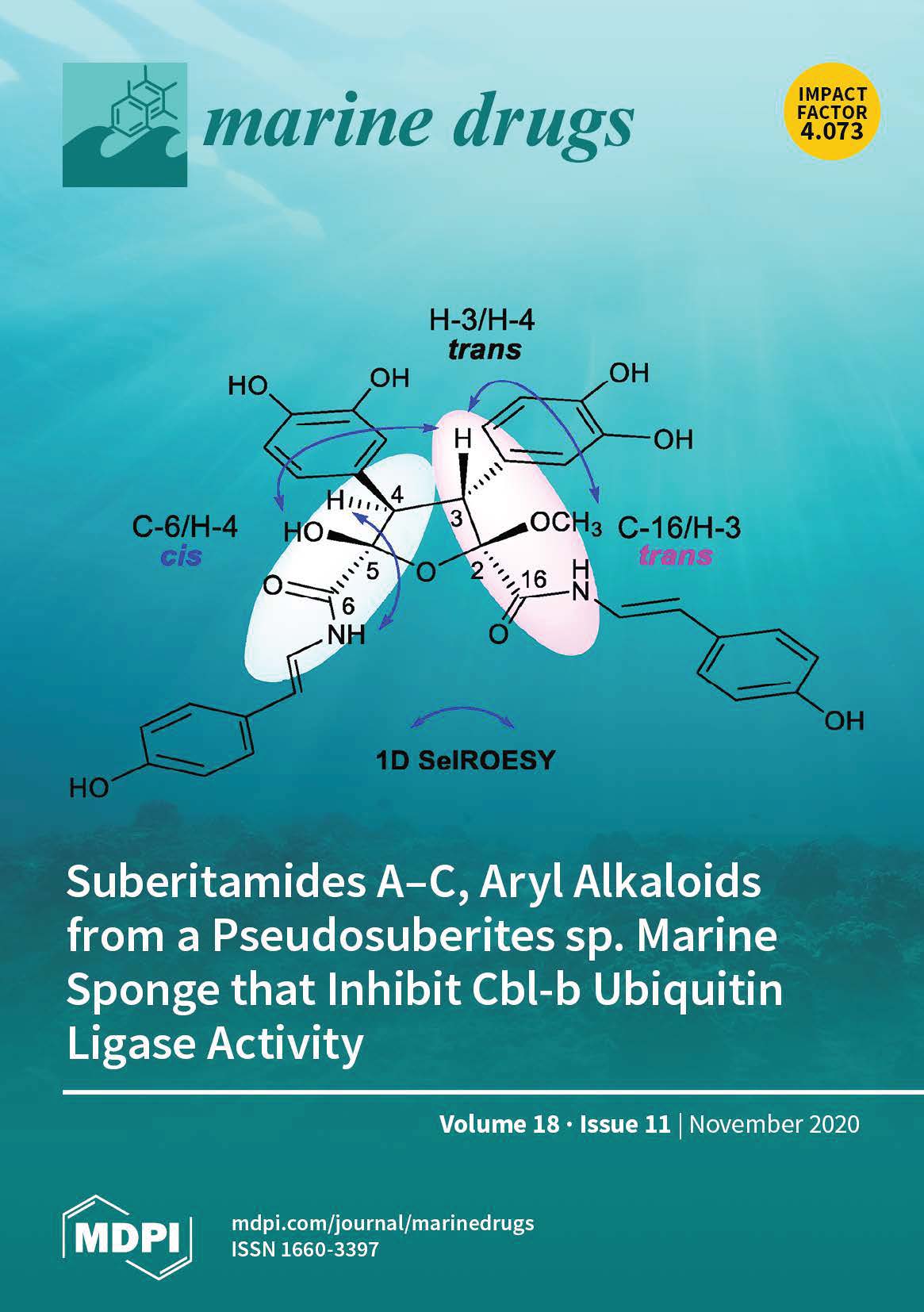
Suberitamides A-C, aryl alkaloids from a Pseudosuberites sp. marine sponge that inhibit Cbl-b ubiquitin ligase activity
Three new aryl alkaloids named suberitamides A-C (1-3), were isolated from an extract of the marine sponge Pseudosuberites sp. collected along the coast of North Carolina. Their planar structures were established by extensive nuclear magnetic resonance (NMR) and mass spectrometry (MS) analysis. To assign the challenging relative configuration of the saturated five-membered ring in suberitamide A (1), a simple and efficient NMR protocol was applied that is based on the analysis of 2- and 3-bond (1)H-(13)C spin-spin coupling constants using a PIP (pure in-phase) HSQMBC (heteronuclear single quantum multiple bond correlation) IPAP (in-phase and anti-phase) experiment. Suberitamides A (1) and B (2) inhibited Cbl-b, an E3 ubiquitin ligase that is an important modulator of immune cell function, with IC50 values of approximately 11 µM.
Kim CK, Wang D, Wilson BAP, Sauri J, Voeller D, Lipkowitz S, O'Keefe BR, Gustafson KR. Suberitamides A-C, aryl alkaloids from a Pseudosuberites sp. marine sponge that inhibit Cbl-b ubiquitin ligase activity. Mar Drugs 18: 2020.

National Cancer Institute (NCI) Program for Natural Products Discovery: Rapid isolation and identification of biologically active natural products from the NCI prefractionated library
An automated, high-capacity, and high-throughput procedure for the rapid isolation and identification of biologically active natural products from a prefractionated library is presented. The semipreparative HPLC method uses 1 mg of the primary hit fraction and produces 22 subfractions in an assay-ready format. Following screening, all active fractions are analyzed by NMR, LCMS, and FTIR, and the active principle structural classes are elucidated. In the proof-of-concept study, we show the processes involved in generating the subfractions, the throughput of the structural elucidation work, as well as the ability to rapidly isolate and identify new and biologically active natural products. Overall, the rapid second-stage purification conserves extract mass, requires much less chemist time, and introduces knowledge of structure early in the isolation workflow.
Grkovic T, Akee RK, Thornburg CC, Trinh SK, Britt JR, Harris MJ, Evans JR, Kang U, Ensel S, Henrich CJ, Gustafson KR, Schneider JP, O'Keefe BR. National Cancer Institute (NCI) Program for Natural Products Discovery: Rapid isolation and identification of biologically active natural products from the NCI prefractionated library. ACS Chem Biol 15: 1104-1114, 2020.

Secondary metabolites from the fungus Dictyosporium sp. and their MALT1 inhibitory activities
Bioassay-guided separation of an extract from a Dictyosporium sp. isolate led to the identification of six new compounds, 1-6, together with five known compounds, 7-11. The structures of the new compounds were primarily established by extensive 1D and 2D NMR experiments. The absolute configurations of compounds 3-6 were determined by comparison of their experimental electronic circular dichroism (ECD) spectra with DFT quantum mechanical calculated ECD spectra. Compounds 3-5 possess novel structural scaffolds, and biochemical studies revealed that oxepinochromenones 1 and 7 inhibited the activity of MALT1 protease.
Tran TD, Wilson BAP, Henrich CJ, Staudt LM, Krumpe LRH, Smith EA, King J, Wendt KL, Stchigel AM, Miller AN, Cichewicz RH, O'Keefe BR, Gustafson KR. Secondary metabolites from the fungus Dictyosporium sp. and their MALT1 inhibitory activities. J Nat Prod 82: 154, 2019.

NCI Program for Natural Product Discovery: A publicly-accessible library of natural product fractions for high-throughput screening
The US National Cancer Institute's (NCI) Natural Product Repository is one of the world's largest, most diverse collections of natural products containing over 230,000 unique extracts derived from plant, marine, and microbial organisms that have been collected from biodiverse regions throughout the world. Importantly, this national resource is available to the research community for the screening of extracts and the isolation of bioactive natural products. However, despite the success of natural products in drug discovery, compatibility issues that make extracts challenging for liquid handling systems, extended timelines that complicate natural product-based drug discovery efforts and the presence of pan-assay interfering compounds have reduced enthusiasm for the high-throughput screening (HTS) of crude natural product extract libraries in targeted assay systems. To address these limitations, the NCI Program for Natural Product Discovery (NPNPD), a newly launched, national program to advance natural product discovery technologies and facilitate the discovery of structurally defined, validated lead molecules ready for translation will create a prefractionated library from over 125,000 natural product extracts with the aim of producing a publicly-accessible, HTS-amenable library of >1,000,000 fractions. This library, representing perhaps the largest accumulation of natural-product based fractions in the world, will be made available free of charge in 384-well plates for screening against all disease states in an effort to reinvigorate natural product-based drug discovery.
Thornburg CC, Britt JR, Evans JR, Akee RK, Whitt JA, Trinh SK, Harris MJ, Thompson JR, Ewing TL, Shipley SM, Grothaus PG, Newman DJ, Schneider JP, Grkovic T, O'Keefe BR. NCI Program for Natural Product Discovery: A publicly-accessible library of natural product fractions for high-throughput screening. ACS Chem Biol 13: 2484-2497, 2018.

Harnessing natural product diversity for fluorophore discovery: Naturally occurring fluorescent hydroxyanthraquinones from the marine crinoid Pterometra venusta
Fluorescent small molecules are important tools in many aspects of modern biology. A two-stage evaluation process involving fluorescence screening and live-cell imaging was developed to facilitate the identification of new fluorescent probes from extracts housed within the NCI Natural Products Repository. To this end, over 2,000 extracts and prefractionated samples were examined, including an extract from the marine crinoid Pterometra venusta. An optically guided evaluation involving stepwise fluorescence screening and live-cell imaging was developed to enable the isolation of fluorescent natural products. These efforts resulted in the isolation of six hydroxyanthraquinone compounds, three of which are new natural products. These purified metabolites were examined for their potential as cellular imaging probes, and they demonstrate that natural product libraries can be a good source of new fluorescent agents.
Singh AJ, Gorka AP, Bokesch HR, Wamiru A, O'Keefe BR, Schnermann MJ, Gustafson KR. Harnessing natural product diversity for fluorophore discovery: Naturally occurring fluorescent hydroxyanthraquinones from the marine crinoid Pterometra venusta. J Nat Prod 81: 2750-2755, 2018.

Unequivocal determination of caulamidines A and B: application and validation of new tools in the structure elucidation tool box
Ambiguities and errors in the structural assignment of organic molecules hinder both drug discovery and total synthesis efforts. Newly described NMR experimental approaches can provide valuable structural details and a complementary means of structure verification. The caulamidines are trihalogenated alkaloids from a marine bryozoan with an unprecedented structural scaffold. Their unique carbon and nitrogen framework was deduced by conventional NMR methods supplemented by new experiments that define 2-bond heteronuclear connectivities, reveal very long-range connectivity data, or visualize the (35,37)Cl isotopic effect on chlorinated carbons. Computer-assisted structural elucidation (CASE) analysis of the spectroscopic data for caulamidine A provided only one viable structural alternative. Anisotropic NMR parameters, specifically residual dipolar coupling and residual chemical shift anisotropy data, were measured for caulamidine A and compared to DFT-calculated values for the proposed structure, the CASE-derived alternative structure, and two energetically feasible stereoisomers. Anisotropy-based NMR experiments provide a global, orthogonal means to verify complex structures free from investigator bias. The anisotropic NMR data were fully consistent with the assigned structure and configuration of caulamidine A. Caulamidine B has the same heterocyclic scaffold as A but a different composition and pattern of halogen substitution. Caulamidines A and B inhibited both wild-type and drug-resistant strains of the malaria parasite Plasmodium falciparum at low micromolar concentrations, yet were nontoxic to human cells.
Milanowski DJ, Oku N, Cartner LK, Bokesch HR, Williamson RT, Sauri J, Liu Y, Blinov KA, Ding Y, Li XC, Ferreira D, Walker LA, Khan S, Davies-Coleman MT, Kelley JA, McMahon JB, Martin GE, Gustafson KR. Unequivocal determination of caulamidines A and B: application and validation of new tools in the structure elucidation tool box. Chemical Science 9: 307-314, 2018.

Enhanced measurement of residual chemical shift anisotropy for small molecule structure elucidation
A method is introduced to measure residual chemical shift anisotropies conveniently and accurately in the mesophase of poly-gamma-(benzyl-l-glutamate). The alignment amplitude is substantially enhanced over common methods which greatly benefits measurements particularly on sp(3) carbons. The approach offers significant improvements in data accuracy and utility for small molecule structure determination.
Liu Y, Cohen RD, Gustafson KR, Martin GE, Williamson RT. Enhanced measurement of residual chemical shift anisotropy for small molecule structure elucidation. Chemical Communications 54: 4254-4257, 2018.

Synthesis and biological assessment of 3,7-dihydroxytropolones
3,7-Dihydroxytropolones (3,7-dHTs) are highly oxygenated troponoids that have been identified as lead compounds for several human diseases. To date, structure-function studies on these molecules have been limited due to a scarcity of synthetic methods for their preparation. New synthetic strategies towards structurally novel 3,7-dHTs would be valuable in further studying their therapeutic potential. Here we describe the successful adaptation of a [5 + 2] oxidopyrilium cycloaddition/ring-opening for 3,7-dHT synthesis, which we apply in the synthesis of a plausible biosynthetic intermediate to the natural products puberulic and puberulonic acid. We have also tested these new compounds in several biological assays related to human immunodeficiency virus (HIV), hepatitis B virus (HBV) and herpes simplex virus (HSV) in order to gain insight into structure-functional analysis related to antiviral troponoid development.
Hirsch DR, Schiavone DV, Berkowitz AJ, Morrison LA, Masaoka T, Wilson JA, Lomonosova E, Zhao H, Patel BS, Datla SH, Hoft SG, Majidi SJ, Pal RK, Gallicchio E, Tang L, Tavis JE, Le Grice SFJ, Beutler JA, Murelli RP. Synthesis and biological assessment of 3,7-dihydroxytropolones. Org Biomol Chem 16: 62-69, 2018.

Five new stilbenes from the stem bark of Artocarpus communis
Five new prenylated stilbenes (1 - 5), along with the known compounds cudraflavone C, trans-4-isopentenyl-3,5,2',4'-terahydroxystilbene, trans-4-(3-methyl-E-but-1-enyl)-3,5,2',4'-tetrahydroxystilbene, pannokin G, cycloartobiloxanthone, artonin P, morusin, artocarpin, artonin E, kuwanon C, artobiloxanthone, and artoindonesianin C (6 - 17) were isolated from the stem bark of the tropical tree Artocarpus communis. The structures were established by NMR spectroscopic analysis, MS studies, and comparison with spectral data reported in the literature.
Chan STS, Popplewell WL, Bokesch HR, McKee TC, Gustafson KR. Five new stilbenes from the stem bark of Artocarpus communis. Natural Products Sciences 24: 266-271, 2018.
A synthetic mirror image of kalata B1 reveals that cyclotide activity is independent of a protein receptor
Featuring a circular, knotted structure and diverse bioactivities, cyclotides are a fascinating family of peptides that have inspired applications in drug design. Most likely evolved to protect plants against pests and herbivores, cyclotides also exhibit anti-cancer, anti-HIV, and hemolytic activities. In all of these activities, cell membranes appear to play an important role. However, the question of whether the activity of cyclotides depends on the recognition of chiral receptors or is primarily modulated by the lipid-bilayer environment has remained unknown. To determine the importance of lipid membranes on the activity of the prototypic cyclotide, kalata B1, we synthesized its all-D enantiomer and assessed its bioactivities. After the all-D enantiomer had been confirmed by (1) H NMR to be the structural mirror image of the native kalata B1, it was tested for anti-HIV activity, cytotoxicity, and hemolytic properties. The all-D peptide is active in these assays, albeit with less efficiency; this reveals that kalata B1 does not require chiral recognition to be active. The lower activity than the native peptide correlates with a lower affinity for phospholipid bilayers in model membranes. These results exclude a chiral receptor mechanism and support the idea that interaction with phospholipid membranes plays a role in the activity of kalata B1. In addition, studies with mixtures of L and D enantiomers of kalata B1 suggested that biological activity depends on peptide oligomerization at the membrane surface, which determines affinity for membranes by modulating the association-dissociation equilibrium
Sando L, Troeira Henriques S, Foley F, Simonsen SM, Daly NL, Hall KN, Gustafson KR, Aguilar MI, Craik DJ. A synthetic mirror image of kalata B1 reveals that cyclotide activity is independent of a protein receptor. Chembiochem 12: 2456-2462, 2011.
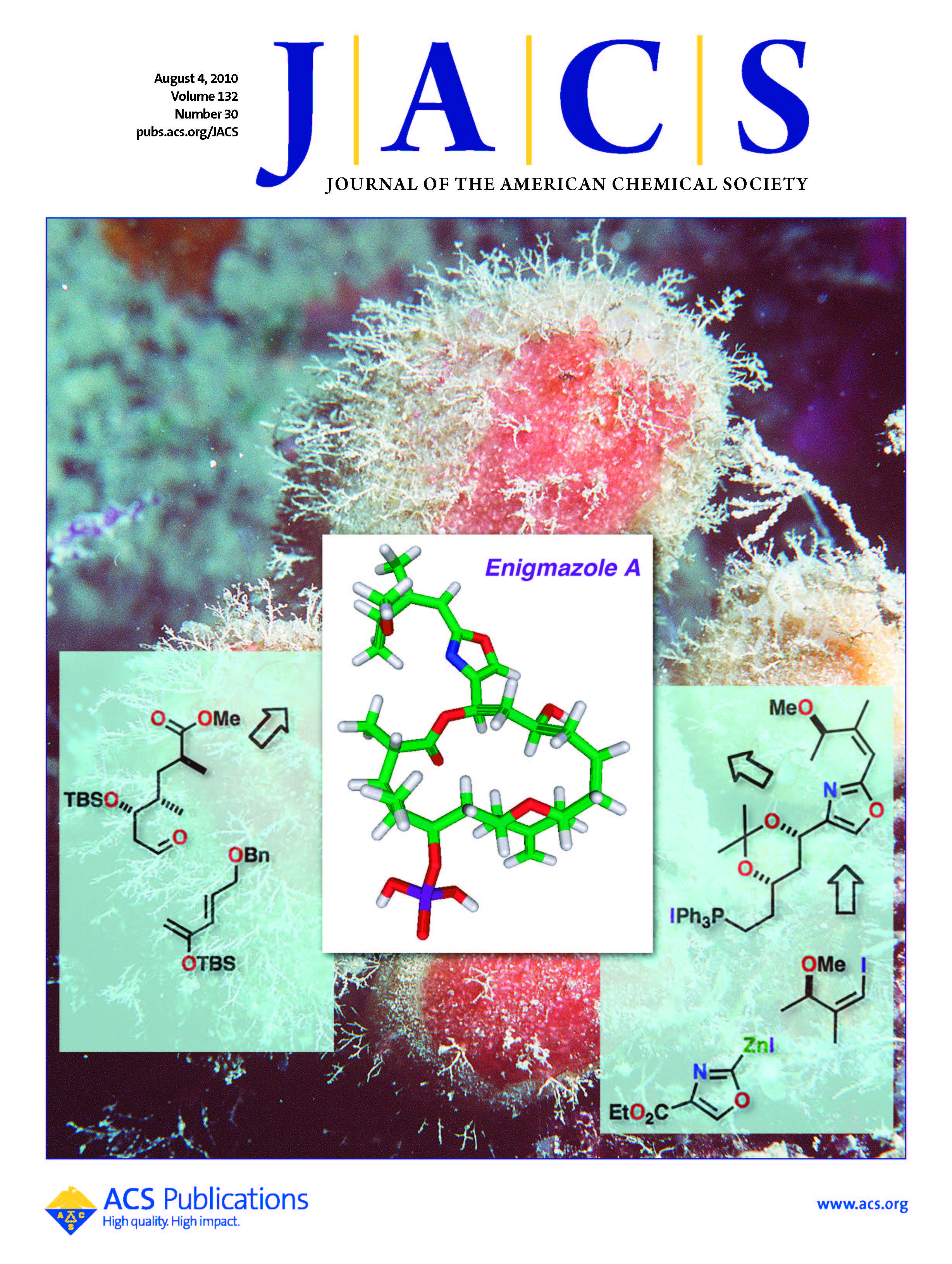
Isolation, structural elucidation, and absolute stereochemistry of enigmazole A, a cytotoxic phosphomacrolide from the Papua New Guinea marine sponge Cinachyrella enigmatica
Enigmazole A (1), a novel phosphate-containing macrolide, was isolated from a Papua New Guinea collection of the marine sponge Cinachyrella enigmatica. The structure of 1, including the absolute stereochemistry at all eight chiral centers, was determined by a combination of spectroscopic analyses and a series of microscale chemical derivatization studies. Compound 1 is comprised of an 18-membered phosphomacrolide that contains an embedded exomethylene-substituted tetrahydropyran ring and an acyclic portion that spans an embedded oxazole moiety. Two additional analogues, 15-O-methylenigmazole A and 13-hydroxy-15-O-methylenigmazole A, were also isolated and assigned. The enigmazoles are the first phosphomacrolides from a marine source and 1 exhibited significant cytotoxicity in the NCI 60-cell line antitumor screen, with a mean GI(50) of 1.7 µM
Oku N, Takada K, Fuller RW, Wilson JA, Peach ML, Pannell LK, McMahon JB, Gustafson KR. Isolation, structural elucidation, and absolute stereochemistry of enigmazole A, a cytotoxic phosphomacrolide from the Papua New Guinea marine sponge Cinachyrella enigmatica. J Am Chem Soc 132: 10278-10285, 2010.

Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high-mannose oligosaccharides
The development of anti-HIV microbicides for either topical or ex vivo use is of considerable interest, mainly due to the difficulties in creating a vaccine that would be active against multiple clades of HIV. Cyanovirin-N (CV-N), an 11kDa protein from the cyanobacterium (blue-green algae) Nostoc ellipsosporum with potent virucidal activity was identified in the search for such antiviral agents. The binding of CV-N to the heavily glycosylated HIV envelope protein gp120 is carbohydrate-dependent. Since previous CV-N – dimannose structures could not fully explain CV-N/oligomannose binding, we determined the crystal structures of recombinant CV-N complexed to Man-9 and a synthetic hexamannoside, at 2.5 Å and 2.4 Å resolution, respectively. CV-N is a three-dimensionally domain-swapped dimer in the crystal structures, with two
primary sites near the hinge region, and two secondary sites on the opposite ends of the dimer. The binding interface is constituted of three stacked a1?2 linked mannose rings for Man-9 and two stacked mannose rings for hexamannoside, with the rest of the saccharide molecules pointing to the solution. These structures show unequivocally the binding geometry of high-mannose sugars to CV-N, permitting a better understanding of carbohydrate binding to this potential new lead for the design of drugs against AIDS.
Botos I, O'Keefe BR, Shenoy SR, Cartner LK, Ratner DM, Seeberger PH, Boyd MR, Wlodawer A. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high-mannose oligosaccharides. J Biol Chem 277: 34336-34342, 2002.

Contact
Contact Info
Center for Cancer Research National Cancer Institute
- Building 576, Room 100D
- Frederick, MD 21702-1201
- 301-846-5391







