Shyam K. Sharan, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 560, Room 32-33
- Frederick, MD 21702-1201
- 301-846-5140
- sharans@mail.nih.gov
RESEARCH SUMMARY
Breast cancer is one of most common malignancies in women. Mutations in BRCA1 and BRCA2 are linked to increased risk of early onset familial breast and ovarian cancers. In an effort to reduce the mortality due to breast cancer through prevention and early diagnosis, sequencing based genetic tests are available to identify BRCA1 and BRCA2 mutation carriers. Such tests have led to identification of several hundreds of sequence variants that are of unknown clinical significance. We are using mouse embryonic stem cells as well as humanized and knock-in mouse models to distinguish between neutral or non-pathogenic and deleterious or pathogenic variants. Dr. Sharan is Deputy Director of the Mouse Cancer Genetics Program (MCGP). Dr. Sharan is also Director of the Center for Advanced Preclinical Research (CAPR) and heads the BRCA Variants Analysis Unit. The BRCA Variants Analysis Unit focuses on functional classification of BRCA2 variants. Click here to access the BRCA2 VARIANT EXPLORER
Areas of Expertise

Shyam K. Sharan, Ph.D.
Research
Each year more than 2 million new cases of breast cancer are diagnosed worldwide. Availability of novel diagnostic tools and targeted therapies together with changes in lifestyle have clearly contributed to a decline in mortality due to breast cancer. While there are many possible risk factors associated with the development of breast cancer, the best-established indicator is the inheritance of breast cancer susceptibility genes such as BRCA1 and BRCA2. Mutations in BRCA1 and BRCA2 are linked to increased risk of early onset familial breast and ovarian cancers with disease penetrance estimated to be 35-80%. In 1997, just a few years after the cloning of BRCA1 and BRCA2, their role in the repair of double strand breaks (DSBs) was uncovered. The proteins encoded by these genes were found to interact with DNA recombinase RAD51, which alluded to their role in homologous recombination (HR). Loss of function mutations in these genes was shown to result in increased genomic instability, which suggested their role as caretakers of the genome.
FUNCTIONAL EVALUATION OF BRCA2 VARIANTS
In an effort to reduce the mortality due to breast cancer through prevention and early diagnosis, sequencing-based genetic tests are available to identify BRCA1 and BRCA2 mutation carriers. Such tests have led to identification of thousands of sequence variants that are of unknown clinical significance due to lack of sufficient epidemiological data that can be used to determine their pathogenicity. The overarching goal of my laboratory over the past two decades has been to understand the functional significance of variants of unknown clinical significance (VUS) identified in BRCA1 and BRCA2 using mouse models as well as mouse embryonic stem cells (mESC). We have undertaken genetic approaches for functional dissection by generating targeted mutations found in breast cancer patients into specific functional domains of BRCA2 in mice. In doing so, we not only aim to functionally classify VUS but also utilize pathogenic variants to uncover new biological functions of these proteins. We believe that functional analysis of pathogenic variants will provide clues to the pathways that are disrupted and lead to tumorigenesis.
We have developed assays to functionally characterize BRCA1 and BRCA2 variants and use our findings, together with other available epidemiological, structural, functional and in silico prediction models to classify the variants as neutral or pathogenic. The extent to which a specific variant complements the lethality of mESC lacking endogenous Brca1 or Brca2 and the known functions of these proteins in DNA repair is used as a surrogate readout to decipher their association with risk of developing the disease. In 2014, due to the clinical relevance of this work, we established the BRCA Variants Analysis Unit in MCGP as a dedicated translational resource for functional classification of BRCA variants. Our current efforts in the unit as well as in my laboratory are primarily focused on BRCA2. We are developing CRISPR-Cas9 based high throughput functional assays for BRCA2 in humanized mESC that lack endogenous Brca2 but carry a single copy of full-length human BRCA2 transgene cloned in a bacterial artificial chromosome.
GENERATION OF BRCA2 MOUSE MODELS
Our laboratory has generated a number of mouse models to assess the effects of BRCA2 variants on growth and development, fertility and tumorigenesis. This approach is especially important for variants that do not severely disrupt the protein function and show mild phenotypic consequences in mESC. We have generated mice with a single amino acid substitution (Gly25Arg) in the PALB2 (Partner and localizer of BRCA2) binding domain of BRCA2 and showed the importance BRCA2-PALB2 interaction in the repair of DSBs by HR as well as protection of stalled replication forks. We have also examined the physiological significance of BRCA2-DSS1 (Deleted in split hand/split foot1) interaction using a knock-in mouse model with a leucine to proline change in codon 2431 (Leu2431Pro, equivalent to human Leu2510Pro), which weakens this interaction. Our functional studies in mESC showed that this interaction is critical for RAD51 recruitment and repair of DSBs by HR. Surprisingly, our functional studies in mice revealed that the interaction is dispensable for repair of DNA replication induced breaks as well as repair of DSBs during meiosis. When bound to BRCA2, DSS1 has been reported to act as a DNA mimic and facilitate RAD51 recruitment by displacing Replication Protein A (RPA) from single-stranded (ss) DNA at DSBs. Our findings suggest that the presence of homologous chromosomes in proximity during early prophase I stage of meiosis may compensate for the defect in BRCA2-DSS1 interaction. This is supported by our observation that tethering a biotinylated homologous DNA to the site of DSBs using streptavidin-fused Cas9 partially rescued the HR defect in mESC expressing this variant.
We are generating new mouse models to functionally characterize another important domain of BRCA2, the BRC domain containing eight BRC repeats spanning over 1100 amino acids. Several of these repeats have been shown to bind to RAD51. Each repeat consists of 35-40 amino acids with an evolutionarily conserved core consisting of hydrophobic residues that facilitate close contact with RAD51. In spite of such an essential function, no cancer causing missense substitution has been identified in this region, which suggests functional redundancy between these repeats. We and others have shown that a single BRC is sufficient for RAD51 recruitment and has partial HR activity in mESC. We are generating mice with a single BRC, repeat 2 or repeat 4, to examine whether the residual HR activity is sufficient for normal physiological functions.
IDENTIFICATION OF BRCA2 GENETIC INTERACTORS
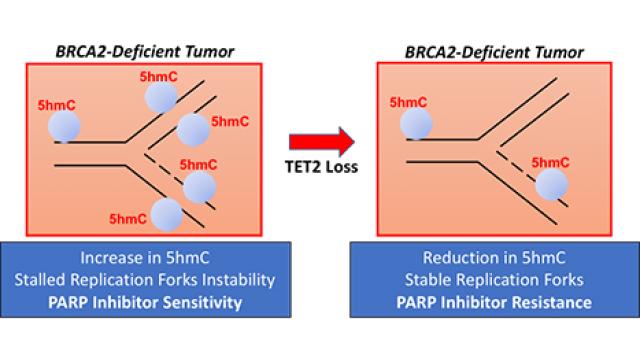
Another major aim of our laboratory is to identify genetic interactors of BRCA2 that may contribute to cell viability and tumorigenesis. This aim is based on the fact that loss of essential genes such as BRCA2 in embryonic stem cells induces cell cycle arrest and apoptosis yet in somatic cells loss of these genes result in tumorigenesis. It is believed that mutations in genes such as TRP53 enables these cells to overcome the growth arrest induced by the loss of such essential genes. We are using mESC to perform genetic screens to identify genes that can contribute to the survival of BRCA2-deficient cells. Because the genome of ES cells is quite stable even after several passages, they represent a powerful and simple yet physiologically relevant system for such screens. More recently, we have initiated a CRISPR/Cas9-based whole genome screen to identify genes that support viability of Brca2ko/ko mESC and may contribute to tumorigenesis. Over the years, we have identified a number of genetic interactors such as GIPC3, PARP1, TET2 and BRE. We have shown GIPC3 to contribute to cell viability via its interaction with APPL1/2, adaptor proteins that regulate multiple signaling pathways. Similarly, BRCC45/BRE contributes to cell viability by recruiting USP7, a deubiquitylase, to stabilize cell cycle regulator CDC25A. More recently, we demonstrated that TET2 loss results in resistance to PARP inhibitors, such as olaparib. Our functional studies revealed that TET2 loss contributed to resistance to PARPi because of its impact on stability of stalled forks. We are currently examining the possibility of modulating TET2 activity that can be used for restoring PARPi sensitivity of resistant tumors in patients.
The long-term objective of my research program is a comprehensive investigation into the biological functions of BRCA2 as well as identification of its genetic interactors. Our work on the genetic interactors will provide insight into initiation of BRCA-mediated tumorigenesis. We anticipate that these studies will result in identification of potential therapeutic targets that can be used to impede or prevent the aberrant path towards tumorigenesis.
Dr. Sharan is also the Director of the Center for Advanced Preclinical Research (CAPR) and head of the BRCA Variants Analysis Unit.
Publications
Characterization of BRCA2 R3052Q variant in mice supports its functional impact as a low-risk variant
Biography

Shyam K. Sharan, Ph.D.
Dr. Sharan obtained his B.Sc. in botany and M.Sc. in genetics from Delhi University, Delhi. In 1994, he obtained his Ph.D. in genetics under Dr. Terry Magnuson at Case Western Reserve University. He then joined Dr. Allan Bradley's laboratory at Baylor College of Medicine as a Howard Hughes Medical Institute Associate. In 1998, Dr. Sharan established the Genetics of Cancer Susceptibility Group in the ABL-Basic Research Program. His group is involved in the functional analysis of tumor suppressor genes in mice. In 1999, Dr. Sharan joined the Center for Cancer Research, NCI. In 2008, Dr. Sharan received the NCI Director's Intramural Innovation Award and in 2009 he received the NIH merit award and the Arthur Flemming Award for his work on functional analysis of BRCA1 and BRCA2. Dr. Sharan is also the Scientific Director of the Center for Advanced Preclinical Research (CAPR).
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Team
Resources
Center for Advanced Preclinical Research
The Center for Advanced Preclinical Research (CAPR) conducts comprehensive preclinical testing of early-stage candidate drugs; Dr. Sharan directs this program. Collaborative teams develop and use reproducible preclinical studies in the development of effective therapeutics and diagnostics for human cancers. Learn more...
Lab Life

Rameesha's farewell lunch (Postbac fellow, 2024-2025), July 31, 2025

Saying goodbye to Aditya Gujalwar, Werner H. Kirsten Student Intern (2024-2025, Mentor: Satheesh Sengodan) May 2025.

Sharan lab holiday lunch, December 2024

Celebrating acceptance of Sounak and BRCA Variant Team's manuscript for publication in Nature, November 12, 2024

Celebrating acceptance of Sounak and BRCA Variant Team's manuscript for publication in Nature, November 12, 2024

Saying good bye to Student Interns Aishwarya Balamurugan and Anushka Sharma (WHK SIP 2023-24). August 2024.

Dillon Dierman's farewell lunch, June 2024

Celebrating acceptance of Satheesh's manuscript for publication in JCI, January 2024

Sharan lab holiday lunch, December 2023

Celebrating acceptance of Arun's Cell Death & Disease, Kajal's Cell Report Method and Sounak’s STAR Protocols and Developmental Cell manuscripts for publication, November 2023

Celebrating publication of Sounak’s paper in PLOS Genetics, November 2023

Saying goodbye to summer interns, Michael and Ishani. August 2023.

Vaish’s Farewell Party, July 2023

Sharan Lab, July 2023
Front row, Left to right: Anushka Sharma, Ishani Biswas, Swati Priya, Vaishnavi Peddibhotla, Teresa Sullivan, Josephine Geh, Arun Mishra, Shyam Sharan
Back row, Left to right: Michael Schifano, Dylan Caylor, Dillon Dierman, Suhas Kharat, Sounak Sahu, and Satheesh Sengodan.
Not in the picture: Mary Albaugh

BRCA Variants Analysis Unit, July 2023
Left to right: Josephine Geh, Ishani Biswas, Eileen Southon, Teresa Sullivan, Shyam Sharan, Dylan Caylor, Sounak Sahu.

Celebrating 25years at NCI Frederick 1998, March 5, 2023

Celebrating 25 years at NCI Frederick, March 5, 2023

Celebrating 25 years at NCI Frederick, March 5, 2023

Sharan Lab Holiday Lunch, December 2022

Celebrating Vaish's Master's thesis defense, November 2022

Kajal's farewell party, September 17, 2022

Celebrating publication of Sounak's paper in Scientific Reports, July 2022.

Virtual Lab meeting, August 2020

Celebrating acceptance of Suhas's manuscript for publication in Science Signaling and Stacey Stauffer's manuscript in Journal of Human Genetics, August 2020

Betty's farewell party, July 31, 2020

Linda's farewell party Feb 29, 2020

Sharan Lab, Holiday lunch 2019

Sharan Lab, November 2019
Front row, left to right: Betty Martin, Teresa Sullivan, Natasha Oberoi, Linda Cleveland.
Back row, left to right: Arun Mishra, Divya Swaminathan, Sakshi Narula, Stacey Stauffer, Eileen Southon, Sounak Sahu, Suhas Kharat, Satheesh Sengodan, Kajal Biswas, Shyam Sharan.

Lab Lunch, August 2019

Sharan Lab, Holiday lunch 2018

Helene's farewell lunch, April 2018

Sharan Lab, 2016
Teresa Sullivan, Chinmayi Pamela, Betty Martin, Stacey Stauffer, Eileen Southon, Susan Lynn North, Susan Reid, Xia Ding, Kajal Biswas, Linda Cleveland, Suhas Kharat, and Vaibhav Singh.

Sharan Lab, 2015
Front row: Betty Martin and Eswary Thirthagiri
Back row, left to right: Stacey Stauffer, Suzanne Hartford, Teresa Sullivan, Kajal Biswas and Xia Ding

Sharan Lab, Holiday lunch 2013

Sharan Lab 2013
Front row, left to right: Kajal Biswas, Eswary Thirthagiri, Melissa Glover, Suzanne Hartford, Xia Ding
Back row, left to right: Susan Lynn North, Laura Brown, Hanna Marks, Betty Martin, Stacey Stauffer and Shyam Sharan

Sharan Lab 2012 (back row, left to right): Kajal Biswas, Susan Lynn North, Suhwan Chang, Seyoun Kim.
front row, left to right: Rajanikant Chittela, Eswary Thirthagiri, Gloria Caballero, Betty Martin, Hamida Thakur
Sharan Lab, April 2011
Front row, left to right: Susan Lynn North, Gloria Caballero, Eswary Thirthagiri, Rajani Kant Chittela
Back row, left to right: Alok Mishra, Xia Ding, Kajal Biswas, Stacey Stauffer, Betty Martin, Suhwan Chang, Shyam Sharan

Sergey Kuznetsov, Postdoctoral Fellow, 2002-2008
Srividya Swaminathan, Postdoc fellow 1999-2004; Yongping Yang, Postdoc fellow 2000-2005; Subha Philip Postdoctoral fellow 2003-2009; Prajakta Varadkar, Postdoctoral Fellow 2003-2005.

First Sharan Lab lunch, 1999
Jennifer Chandler, Srividya Swaminathan, Yong-Tae Ahn and his son, Shyam Sharan and Shikha Sharan
Our work featured on the cover of Developmental Cell January 22, 2024.
Sahu S, Sahoo S, Sullivan T, O'Sullivan TN, Turan S, Albaugh ME, Burkett S, Tran B, Salomon DS, Kozlov SV, Koehler KR, Jolly MK, Sharan SK.Spatiotemporal modulation of growth factors directs the generation of multilineage mouse embryonic stem cell-derived mammary organoids. Dev Cell. 2024 Jan 22;59(2):175-186.e8.

Our work featured on the cover of Science Signaling, August 20, 2020.
Kharat SS, Ding X, Swaminathan D, Suresh A, Singh M, Sengodan SK, Burkett S, Marks H, Pamala C, He Y, Fox SD, Buehler EC, Muegge K, Martin SE, Sharan SK. Degradation of 5hmC-marked stalled replication forks by APE1 causes genomic instability. Sci Signal. 2020 Aug 18;13(645):eaba8091. doi: 10.1126/scisignal.aba8091.