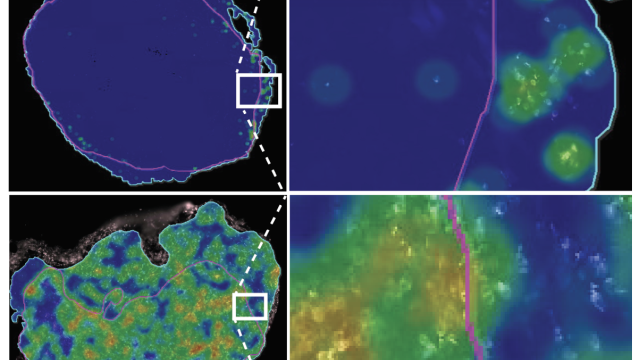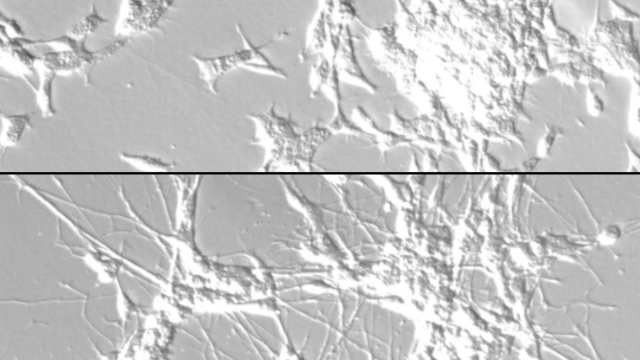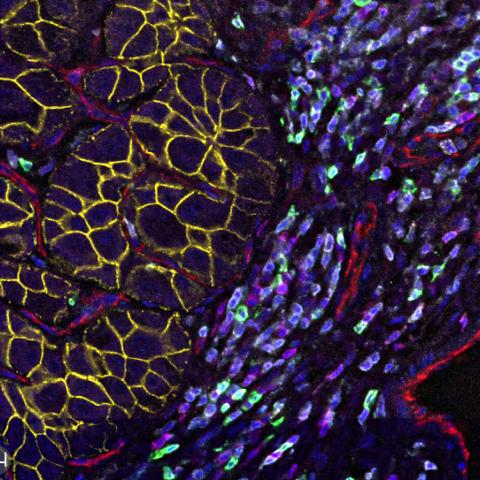
Tumor cell community in a hepatocellular carcinoma, a type of liver cancer.
Image credit: Noemi Kedei and Evan Maestri
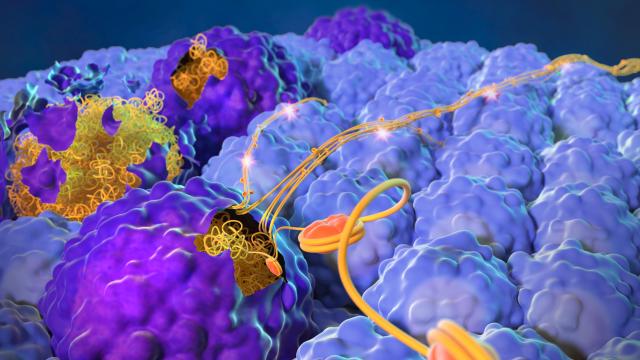
CCR researchers have uncovered stable molecular networks that cells within liver tumors use to communicate with immune cells in the tumor microenvironments. Using single-cell RNA sequencing analyses on samples from 44 liver cancer patients, the investigators identified stable ligand-receptor pairs, which are messenger molecules specifically bound to partner proteins, that tumor cells use to communicate with other cells such as macrophages. The macrophages can engulf and destroy foreign substances, thereby affecting the viability of a tumor.
The findings by senior author Xin Wei Wang, Ph.D., Deputy Chief of the Laboratory of Human Carcinogenesis and co-director of the Liver Cancer Program, and colleagues appeared December 7 in Nature Communications. They found that these networks are consistent among the variety of cancer cells within each tumor, though the networks can vary among patients.
“We identified at least two different communication signals that may be important to how macrophages sense or interact with tumor cells,” Wang says. “Somehow, these receptors are connected to each other. These sensing mechanisms make the tumor more aggressive, possibly reprogramming the macrophages to be functionally different or more active.”
The genetic variation between cancer cells within a single tumor makes it difficult to treat the cells with broad-based therapies or genetically-targeted treatments, as the features and available targets of cells in one region can vary from those of cells in another region. This variation can also give rise to sampling bias, in which a single biopsy that clinicians use to determine the biology of a tumor and how to treat it may not be representative of the whole tumor environment.
Single-cell RNA sequencing can be used to identify a constant feature amid the changing environment within liver cancer tumors, as these tumors are less likely to be driven by single, dominant mutations than other forms of cancer. Use of this sequencing might lead to a reliable treatment target in a liver cancer patient.
“By using this method, we may be able to profile liver cancer patients using a single biopsy,” says Lichun Ma, Ph.D., a Stadtman Investigator in the Cancer Data Science Laboratory affiliated with the Liver Cancer Program and first author on the paper. “These ligand-receptor networks might help to open a path for therapeutic exploration because we could target and block the communications between tumor cells and tumor-associated macrophages.”
Future treatments might involve patients being classified into subgroups based on similarities among the communication networks in their tumor microenvironment, she says.
The next steps for the researchers will involve exploring the functional consequences of the interactions between the two ligand-receptor pairs in both cell cultures and animal models of liver cancer, Ma says. They are also planning to investigate how these networks may change in response to therapy in clinical trials as liver cancer cells evolve to develop resistance to treatments.