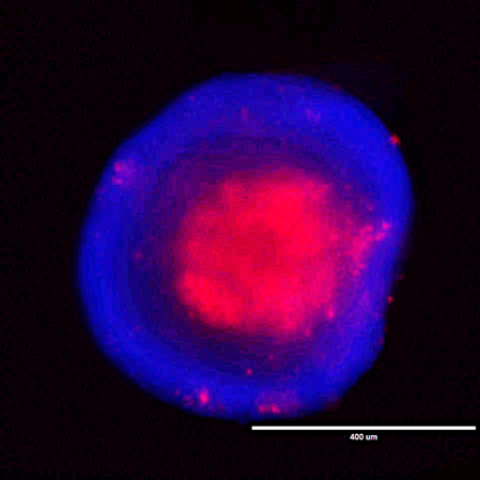
HCT116 (KRAS-mutant) colorectal cancer cell spheroid. The blue shows cell nuclear staining and red shows dead cell staining.
Photo courtesy of Chih-Shia Lee.
Researchers in CCR examined nearly 500 combinations of genes thought to be critical for maintaining KRAS-mutant cancer cell survival over normal cells. They found that the combination of BRAF, CRAF and ATG7 is responsible for favoring the KRAS-mutant cancer cells over normal cells. Furthermore, two pathways, MAPK and autophagy, work together to keep KRAS-driven cancer cells alive.
KRAS is known to be the driver behind colorectal, pancreatic and other solid tumors. But developing effective targeted therapy to directly inhibit the activity of KRAS oncoproteins has proven to be elusive. This finding, however, may bring researchers a little closer to realizing this goal. The study was published in Proceedings of the National Academy of Sciences (PNAS) on February 1, 2019.
Working in the Laboratory of Cancer Biology and Genetics, Chih-Shia Lee, Ph.D., Research Fellow, along with Investigator Ji Luo, Ph.D., and his team, analyzed about 500 combinations of from one-to-six different genes in KRAS-mutant colorectal and pancreatic cancer and normal cell lines. Their goal was to determine which combinations facilitated KRAS-mutant cancer growth.
“It turns out that BRAF, CRAF and ATG7 were the golden combination,” explained Lee. “BRAF and CRAF are both in the RAF family, and they are part of the MAPK pathway, which drives cell proliferation and growth. ATG7 is part of the autophagy pathway, which is responsible for degrading cell components in order to be self-sufficient when nutrients are limited. This finding suggests that these three proteins need to be inhibited to deactivate KRAS-driven cancers while sparing normal cells.”
The finding also helps shed light on why it has been so difficult to treat these types of cancer. Not only are they driven by two pathways working cooperatively, but the KRAS oncoprotein is quite small and round. Lee noted that it also doesn’t have deep pockets, making it difficult for targeted therapy that uses small molecules to find a place to bind within the protein’s critical structure.
A few inhibitors for BRAF, however, are available, and they work quite well for BRAF-driven melanoma. But they have proven to ineffective for KRAS-driven tumors. “The reason BRAF inhibitors don’t work is that while inhibiting the activity of BRAF, they paradoxically activate the MAPK signaling pathway, keeping the cancer robust,” said Lee. “This is yet another reason why KRAS-driven cancer is hard to beat.”
This study reveals the previously underappreciated complexity of the signaling network of the KRAS oncogene. Although work remains to be done, the research does suggest potential target combinations for more effective therapeutic interventions.