John Brognard, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 560, Room 22-31
- Frederick, MD 21702
- 301-846-1163
- 301-846-1666
- john.brognard@nih.gov
RESEARCH SUMMARY
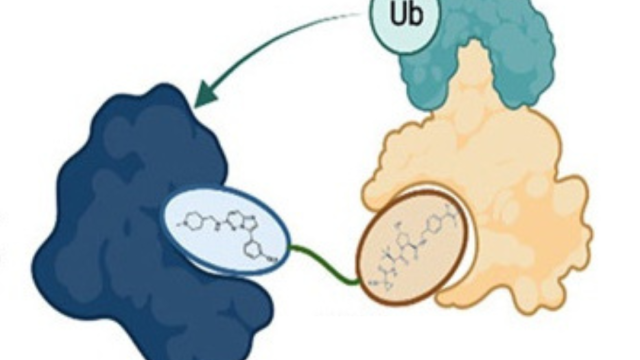
My lab studies cancer-associated kinases in the unexplored kinome, using bioinformatics and functional genomic approaches, to understand the molecular mechanisms by which these kinases promote tumorigenesis. With in vivo patient-derived xenograft (PDX) mouse models, we will translate these findings to the clinic by developing novel compounds, such as PROTACs (proteolysis targeting chimeras), or by repurposing existing drugs to target these kinase drivers of cancer. The goal is to provide a platform for transformational research to identify novel druggable drivers to benefit cancer patients without durable treatment options.
Areas of Expertise

John Brognard, Ph.D.
Research
Cancer genomic sequencing has increased our understanding of the temporal and spatial genetic alterations that lead to tumorigenesis, thereby enabling the development of targeted therapies leading to durable and less toxic responses in patients. Mutations that alter kinase activity or pathways involving kinases are common in cancer. However, most research efforts have focused on ~50 kinases among the 538 kinases present in the human kinome. Data from siRNA screens and cancer genomic studies indicate that at least 300 of the unexplored kinases are implicated in cancer and harbor putative driver mutations.
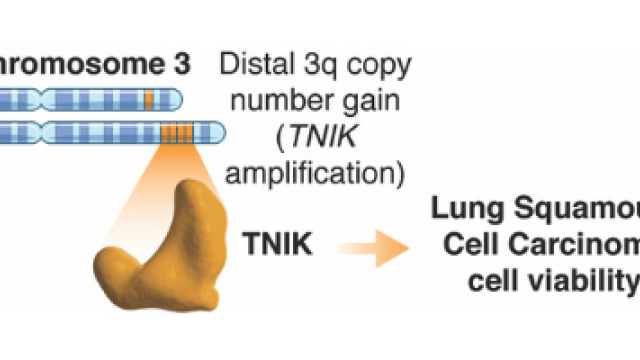
Our research involves defining novel kinase drivers in 3q amplicon-positive squamous cell carcinomas and the mechanisms by which these kinases promote cancer. Our previous work has led us to focus on leucine zipper bearing kinase (LZK, also known as MAP3K13) and TRAF2- and NCK-interacting kinase (TNIK) as therapeutic targets in head and neck squamous cell carcinomas (HNSCCs) and lung squamous cell carcinomas (LSCCs). In addition, we use motif-driven bioinformatic screens across many diverse cancers (pan-cancer) to define novel oncogenic and tumor-suppressing kinases. Combined, these projects will aid in identification of novel kinase drivers of cancer.
We apply diverse approaches to identify cancer-associated kinases and to investigate the signaling pathways regulated by cancer-associated kinases to promote tumorigenesis. These approaches include reverse phase protein arrays (RPPAs) to determine the phosphorylation levels and abundance of proteins, whole-exome sequencing combined with RNAseq to identify genetic mutations in expressed genes, whole-genome sequencing to define copy number alterations, phospho-antibody arrays to globally evaluate activation of signaling pathways, siRNA screens to identify cancer dependencies, molecular dynamics-based modeling of mutations in kinases to provide insight into functional consequences of the mutations and molecular sites for potential therapeutic intervention, and motif-driven, pan-cancer analyses to assess at the kinome level the function of tumor suppressing kinases in cancer. Collectively, this research has and will continue to identify new cancer drivers, targets for therapeutic intervention, and mechanisms of tumorigenesis.
Publications
- Bibliography Link
- View Dr. Brognard's PubMed Summary.
The SRC family kinase inhibitor NXP900 demonstrates potent antitumor activity in squamous cell carcinomas
PIM1 targeted degradation prevents the emergence of chemoresistance in prostate cancer
TNIK is a therapeutic target in Lung Squamous Cell Carcinoma and regulates FAK activation through Merlin
The protein kinase MAP3K19 phosphorylates MAP2Ks and thereby activates ERK and JNK kinases and increases viability of KRAS-mutant lung cancer cells
Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor
Biography

John Brognard, Ph.D.
John received a B.Sc. degree in chemistry from James Madison University (Harrisonburg, Virginia), followed by a M.Sc. degree in biotechnology from Johns Hopkins University (Baltimore, Maryland). He obtained his Ph.D. from University of California, San Diego (UCSD) in Dr. Alexandra Newton’s laboratory, where he discovered a novel class of phosphatases (PHLPP1 and PHLPP2) that act directly on Akt and PKC to decrease signaling through these pathways. Lastly, John trained as a postdoctoral fellow in Dr. Tony Hunter’s laboratory at the Salk Institute. Utilizing bioinformatic tools they screened cancer genomes to identify novel kinases implicated in cancer by the presence of functional somatic mutations. He joined the CRUK Manchester Institute as a group leader in September of 2010 and the lab was focused on identifying mechanisms to promote lung tumorigenesis. John then moved to the Laboratory of Cell and Developmental Signaling at the NCI as a Earl Stadtman Investigator in the summer of 2016. His research focuses on defining novel enzymes that act to suppress or promote tumorigenesis and, in some cases, can serve as novel targets for therapeutic intervention. He has received numerous awards including the Presidential Early Career Award for Scientists and Engineers (PECASE), the CRUK Future Leader in Cancer Research Award, the NCI Director’s award, an NCI Milestone award, and the Inaugural UCSD Pharmacology Department Outstanding Alumnus Award. He is a member of the Faculty of 1000 and serves on the board of reviewing editors at Science Signaling. He received tenure from NIH in 2022.
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.