Yun-Xing Wang, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Bldg. 538/Room 128
- Frederick, MD 21702-1201
- 301-846-5985
- wangyunx@mail.nih.gov
RESEARCH SUMMARY
Yun-Xing Wang is a structural biologist who pioneered the combined use of NMR spectroscopy, small angle X-ray scattering (SAXS), diffraction, atomic force microscopy (AFM) and other biophysical methods to study RNA structural biology. His laboratory research places emphases on solving long-standing important structural biology questions and uses the gained knowledge to develop diagnostic and therapeutic agents for cancer and HIV/AIDS. His laboratory also invented the first RNA labeling “machine”.
Visit the SAXS Facility for more information about this new resource.
For useful resources, visit the Protein-Nucleic Acid Interactions wiki site.
Areas of Expertise

Yun-Xing Wang, Ph.D.
Research
We study the RNA structural biology using biophysical methods, including NMR spectroscopy, SAXS, X-ray diffraction and atomic force microscopy, to determine RNA three-dimensional structures or topological folds, understand RNA function, and provide insight into fundamental mechanisms of RNA biological functions.
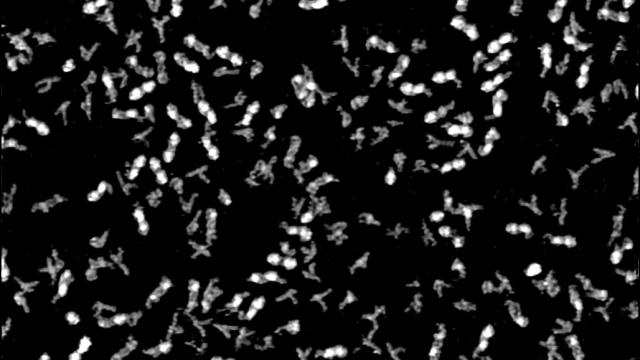
RNA function is coded in its three-dimensional structures and dynamics. To characterize RNA in four-dimensional space, we have resorted to new technologies that allow us to study ever more challenging problems. For this reason, our laboratory has developed Position-selective Labeling of RNA (PLOR) technology, which can be applied for structural biology studies, imaging and disease detection. We have also developed a new approach using X-ray free electron laser (XFEL) and time-resolved nanocrystallography to study RNA structures and dynamics. Recently, we developed the algorithm to recapitulate the 3D topological structure of individual RNA molecules in solution from AFM topographic particle images, which makes it possible to study RNA conformational heterogeneity under physiologically relevant solution conditions, as opposed to in restricted or confined crystal lattices or under artificially high [Mg2+].
Publications
- Bibliography Link
- View Dr. Wang's PubMed Summary.
Determining structures of RNA conformers using AFM and deep neural networks
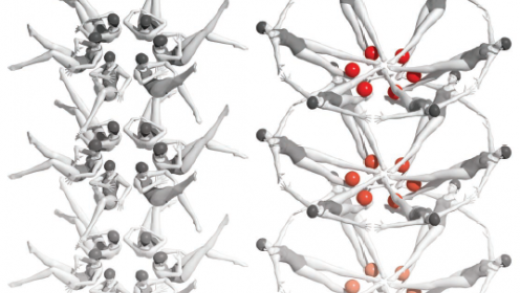
The conformational space of RNase P RNA in solution
Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography
Synthesis and Applications of RNAs With Position-Selective Labelling and Mosaic Composition
An unusual topological structure of the HIV-1 Rev response element
Biography

Yun-Xing Wang, Ph.D.
Dr. Wang conducted his graduate work on structure determination of fragments of 23S rRNA using NMR spectroscopy and UV-melting experiments in Professor David E. Draper's lab of the Johns Hopkins University. Dr. Wang received his Ph.D. in October 1994 from the Johns Hopkins University. From 1994 to 2000 he was an NIH postdoctoral later a research fellow in Dr. Dennis Torchia's laboratory where he studied the structure, hydration dynamics of HIV-1 protease in complex with inhibitors and elucidated the 3D structure and a new function of the antitumor/anti-HIV protein MAP30. In the late 2000 he then joined the Structural Biophysics Laboratory of NCI. Using high field NMR spectroscopy and other biophysical and biochemical methods, his group studies the functional structural biology of RNAs and proteins.
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.