Michael Bustin, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 37, Room 3122
- Bethesda, MD 20892-4255
- 240-760-6867
- bustinm@mail.nih.gov
RESEARCH SUMMARY
Dr. Bustin studies the role of chromosomal proteins in chromatin function, epigenetic regulation, development and disease.
Areas of Expertise

Michael Bustin, Ph.D.
Research
Chromosomal Proteins and Chromatin Function
Precise and specific interactions between chromosomal proteins and the chromatin fiber play a key role in epigenetic regulation, transcription, replication and DNA repair and therefore affect the orderly progression of biological processes such as development and differentiation. Numerous diseases, including cancer, are associated with changes in chromatin structure and function. Dr. Bustin’s research focused on the molecular mechanisms whereby nucleosome-binding architectural proteins such as histone H1 and HMGs affect the structure and function of chromatin and play a role in establishing the cellular phenotype. He was among the first to provide information on a histone amino acid sequence and to study the structure and heterogeneity of histone H1 variants. He produced antibodies against purified histones and HMG proteins and pioneered the use of immunochemical approaches to study chromatin structure and function. Subsequent research in his laboratory focused on the HMGNs, a protein family ubiquitously present in the nuclei of vertebrate cells. His laboratory discovered new HMGN protein variants, cloned and mapped the HMGN genes and generated genetically altered mice for all known HMGN proteins. Using these reagents his laboratory found that HMGN and histone H1 bind dynamically to chromatin and compete for nucleosome binding sites. He demonstrated that HMGN proteins bind preferentially to chromatin regulatory sites, stabilize cell identity, modulate epigenetic regulatory processes and play a role in determining the cellular phenotype. His studies highlight the importance of nucleosome-binding architectural proteins in epigenetic regulation. As an NIH Scientist Emeritus, Dr. Bustin continues his studies on the role of chromosomal proteins in epigenetic regulatory processes that affect chromatin functions and play a role in development and disease.
For more details about Dr. Bustin's research and for representative results, see HMG Proteins and Gallery on this website.
Publications
- Bibliography Link
- View Dr. Bustin's PubMed Summary.
Epigenetic regulation of white adipose tissue plasticity and energy metabolism by nucleosome binding HMGN proteins
Binding of HMGN proteins to cell specific enhancers stabilizes cell identity
Interplay between H1 and HMGN epigenetically regulates OLIG1&2 expression and oligodendrocyte differentiation
Nongenetic functions of the genome
Determinants of histone H1 mobility and chromatin binding in living cells
Biography

Michael Bustin, Ph.D.
Michael Bustin received his Ph.D. from the University at California, Berkeley and did postdoctoral work in the area of protein chemistry in the laboratory of the Nobel laureates, Drs. S. Moore and W. Stein, at the Rockefeller University in New York, and in the area of immunochemistry at the Weizmann Institute of Science, Israel, where he produced antibodies to histones and pioneered their use for studies on chromatin structure and function. Dr. Bustin joined the NIH in 1975. He is the recipient of a Janett and Samuel Lubell Prize from the Weizmann Institute (1975), a 1989 NCI award, a 1993 Tosse-Preis fur Kinderrheumatologie from the German Rheumatology Society, a 1997 Humboldt Research Award, and a 2007 Jacob and Lena Foundation professorship at Hebrew University. Dr. Bustin served as an adjunct professor at Georgetown University (1984-1990) and at Tel Aviv University. His research interests center on the role of chromosomal proteins in chromatin function, gene expression, development and disease.
Dr. Bustin retired in 2022 and is now an NIH Scientist Emeritus.
Covers
ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming
The cover image depicts the role of mitochondrial ADHFE1 in D-2-hydroxyglutarate production, highlighting the contributions of the enzyme and oncometabolite to breast cancer progression.
Metabolic reprogramming in breast tumors is linked to increases in putative oncogenic metabolites that may contribute to malignant transformation. We previously showed that accumulation of the oncometabolite, 2-hydroxyglutarate (2HG), in breast tumors was associated with MYC signaling, but not with isocitrate dehydrogenase (IDH) mutations, suggesting a distinct mechanism for increased 2HG in breast cancer. Here, we determined that D-2HG is the predominant enantiomer in human breast tumors and show that the D-2HG–producing mitochondrial enzyme, alcohol dehydrogenase, iron-containing protein 1 (ADHFE1), is a breast cancer oncogene that decreases patient survival. We found that MYC upregulates ADHFE1 through changes in iron metabolism while coexpression of both ADHFE1 and MYC strongly enhanced orthotopic tumor growth in MCF7 cells. Moreover, ADHFE1 promoted metabolic reprogramming with increased formation of D-2HG and reactive oxygen, a reductive glutamine metabolism, and modifications of the epigenetic landscape, leading to cellular dedifferentiation, enhanced mesenchymal transition, and phenocopying alterations that occur with high D-2HG levels in cancer cells with IDH mutations. Together, our data support the hypothesis that ADHFE1 and MYC signaling contribute to D-2HG accumulation in breast tumors and show that D-2HG is an oncogenic metabolite and potential driver of disease progression.
ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming. Mishra P, Tang W, Putluri V, Dorsey TH, Jin F, Wang F, Zhu D, Amable L, Deng T, Zhang S, Killian JK, Wang Y, Minas TZ, Yfantis HG, Lee DH, Sreekumar A, Bustin M, Liu W, Putluri N, and Ambs S. J Clin Invest. 128(1):323-340, 2018.

Special Issue: High Mobility Group (HMG Proteins): From Chromatin Formation to Cellular Phenotype
High mobility group proteins. Bustin M. Biochim Biophys Acta. 2010 Jan-Feb;1799(1-2):1-2.

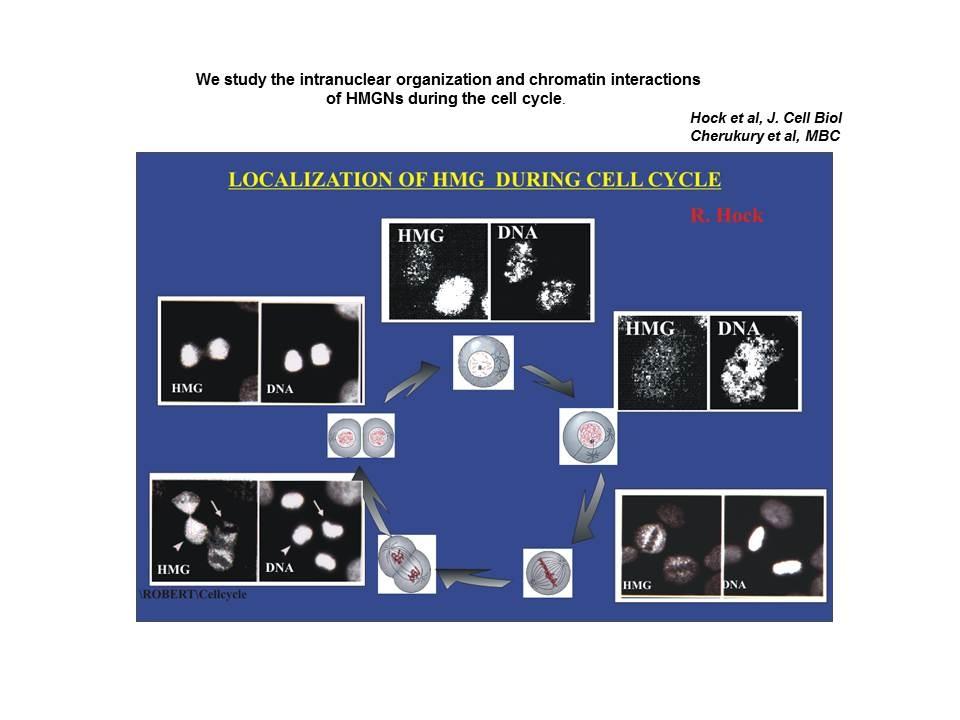
Differential localization of nucleosomal binding proteins during the cell cycle
Shown are confocal images of HeLa cells doubly labeled with antibody to Histone H1 and antibody to HMGN2.
Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Prymakowska-Bosak M, Misteli T, Herrera JE, Shirakawa H, Birger Y, Garfield S, Bustin M. Mol Cell Biol. 2001 Aug;21(15):5169-78.
HMG Proteins
Background
High mobility group (HMG) proteins are ubiquitous nuclear proteins that regulate and facilitate various DNA-related activities such as transcription, replication, recombination and repair. HMGs bind to DNA and chromatin and act as "architectural elements" that induce both short- and long-range changes in the structure of their binding sites. They affect the activities of various regulatory molecules, including hormone receptors, p53, the RAG proteins involved in V(D)J recombination, the homeotic protein HOXD9 of HIV integrase, and several transcription factors. The functional motifs of the ubiquitous HMG proteins are widespread and found in the DNA binding domains of numerous regulatory proteins.
HMG proteins may play a significant role in human disorders. Disruptions and rearrangements in the genes coding for some of the HMG proteins are associated with the etiology of common benign tumors such as uterine leiomyomas, endometrial polyps, and lipomas. HMG-1 and HMG-2 proteins are implicated in the mechanism of action of the anti-cancer drug cisplatinum. In addition, antibodies to HMG proteins are found in patients suffering from autoimmune diseases.
Thus, by modifying the architecture of DNA and chromatin the HMG proteins may contribute to the regulation of various processes that ultimately affect the cellular phenotype. Studies on the structure and function of these proteins may provide insights into the molecular mechanisms underlying the etiology of certain diseases.
Nomenclature
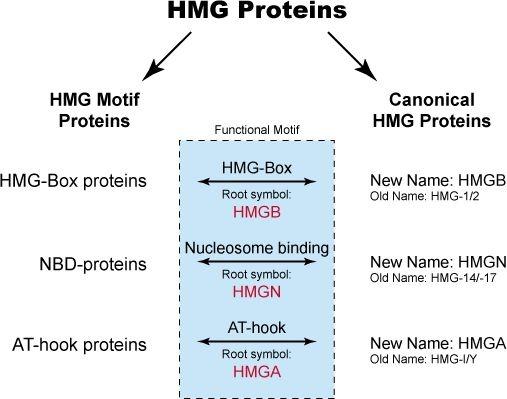
The HMG proteins were originally isolated from mammalian cells and arbitrarily classed as a specific type of nonhistones based on the observation that these proteins are present in all mammalian and many vertebrate cells, that they share certain physical properties, and that they are associated with isolated chromatin. They were originally subdivided into three groups: HMG-1/-2, HMG-14/-17 and HMG-I/Y. These proteins are considered the canonical HMG proteins. Subsequent studies have revealed that the functional motifs characteristic of each of the canonical HMG proteins are widespread among nuclear proteins. Proteins containing any of the functional motifs of the canonical HMG proteins are known as HMG-motif proteins. In fact, the canonical HMG proteins can be considered to be a subclass of the HMG-motif proteins. The HMG-motif proteins are now subdivided into three superfamilies:
- HMGB (formerly HMG-1/-2)
- HMGN (formerly HMG-14/-17)
- HMGA (formerly HMG-I/Y/C)
Each HMG family has a characteristic functional sequence motif. The functional motif of the HMGB family is named "HMG-box", that of the HMGN family is named “nucleosomal binding domain”, and that of the HMGA family is named “AT-hook”. Proteins containing any of these functional motifs embedded in their sequence are known as HMG-motif proteins. The interrelationship between the various HMG proteins is diagrammed below.
HMG Chromosomal Proteins Homepage
For more detailed information and a list of HMG proteins, visit the HMG Chromosomal Proteins Homepage.
References
The HMG Chromosomal Proteins, E.W. Johns (editor). Academic Press, 1982.
Gallery
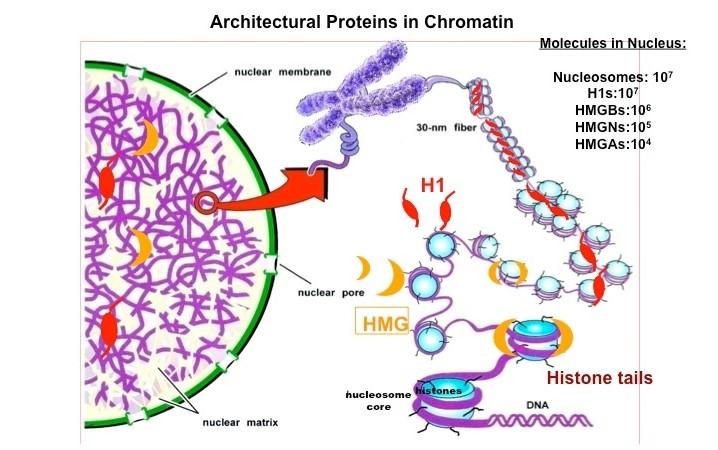
Chromatin Architectural Proteins. Chromatin architectural proteins are structural proteins, devoid of enzymatic activity, that bind to nucleosomes, alter the architecture of their binding sites, and affect chromatin function. The linker histone H1 and members of the HMG superfamily are architectural proteins.
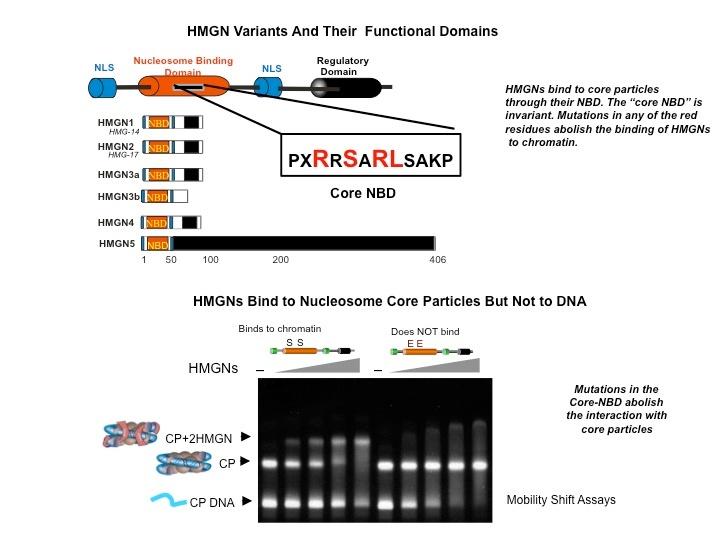
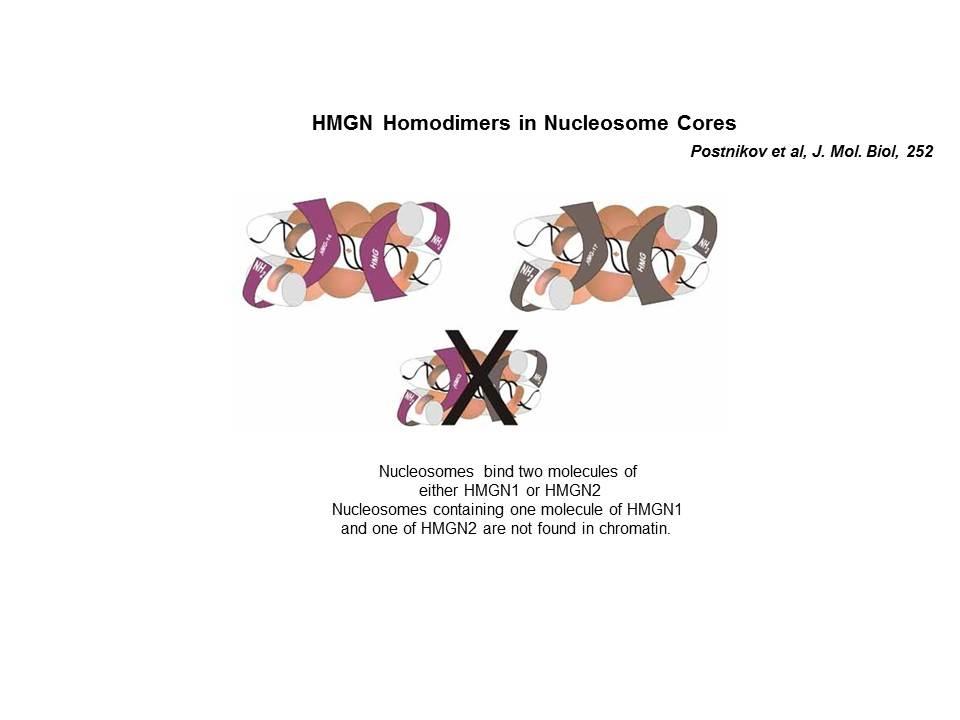
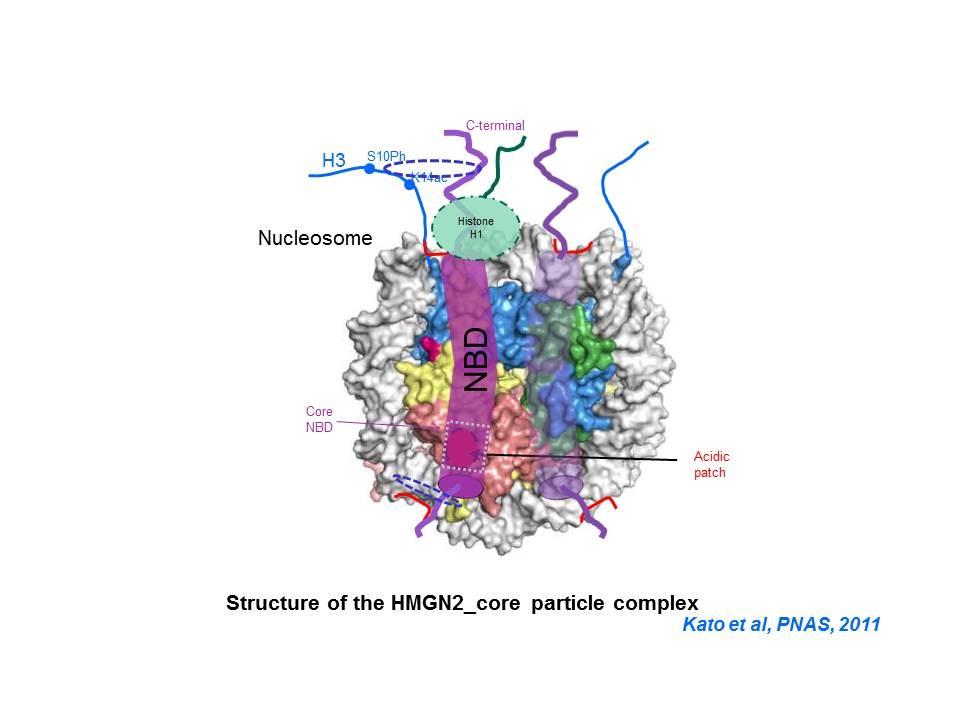
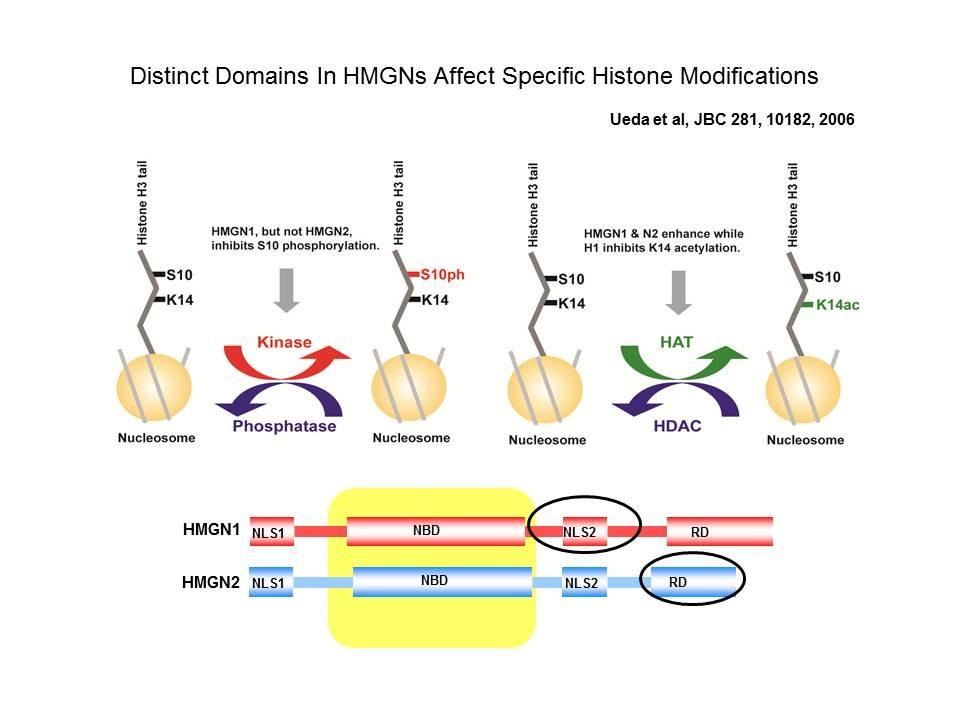
Top: HMGN Variants and Their Functional Domains. HMGNs bind to core particles through their NBD. The "core NBD" is invariant. Mutations in any of the red residues abolish the binding of HGMNs to chromatin. Bottom: HMGNs Bind to Nucleosome Core Particles But Not to DNA. Mutations in the Core-NBD abolish the interaction with core particles.
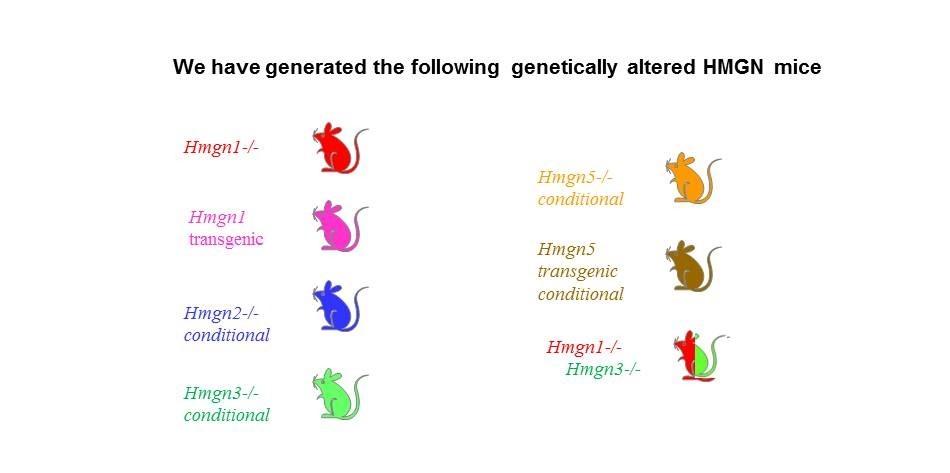
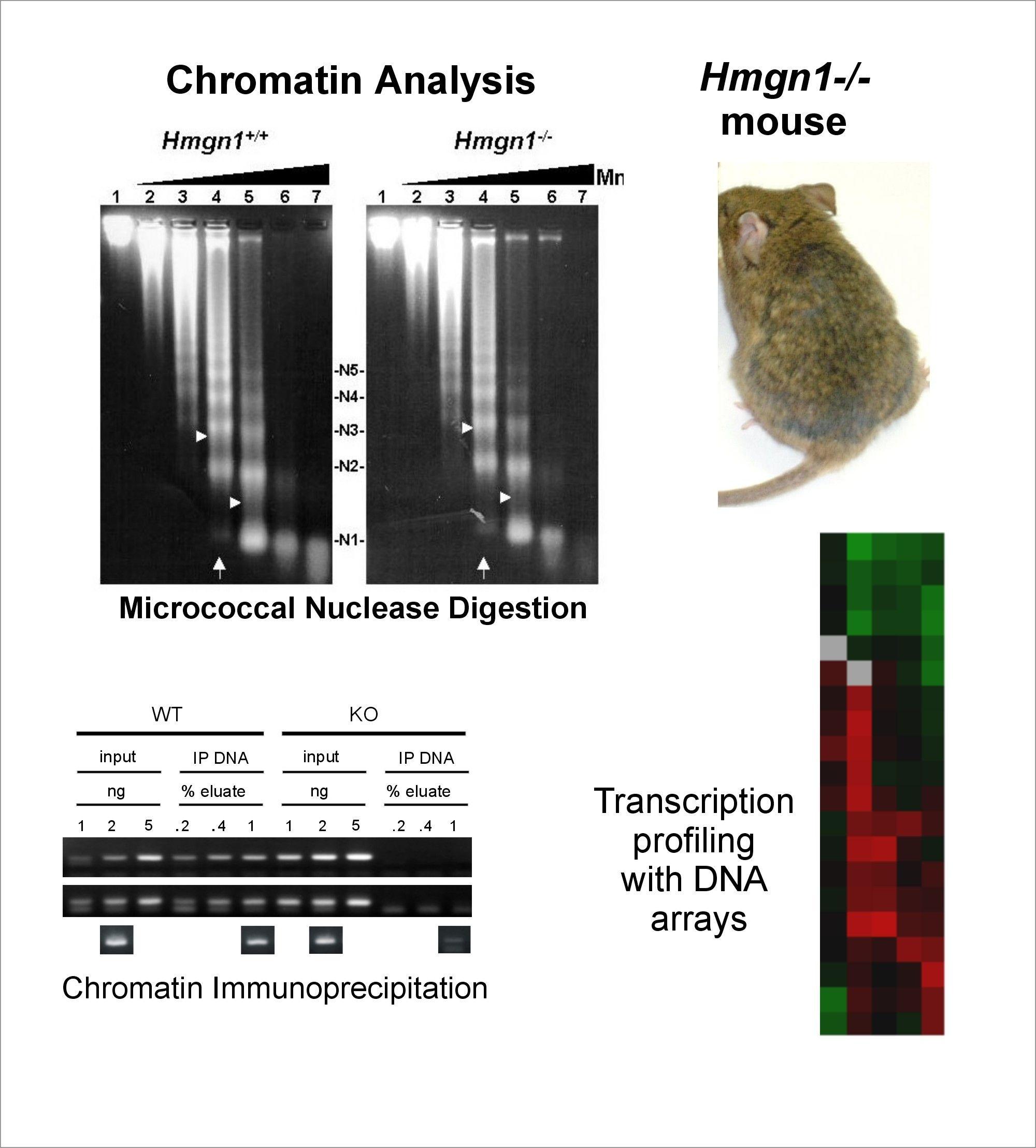
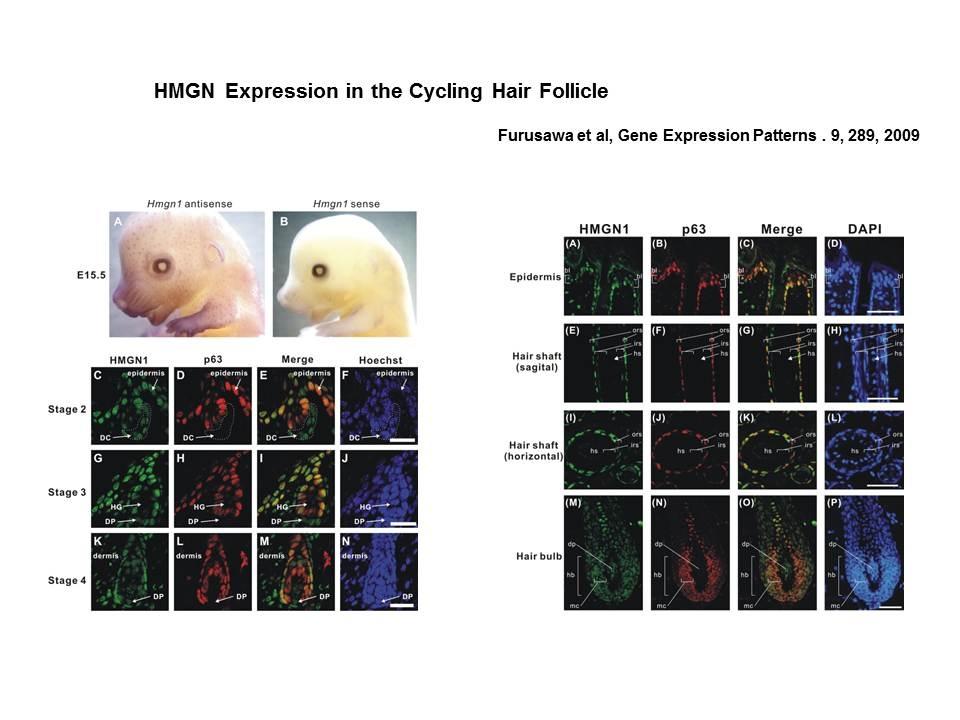
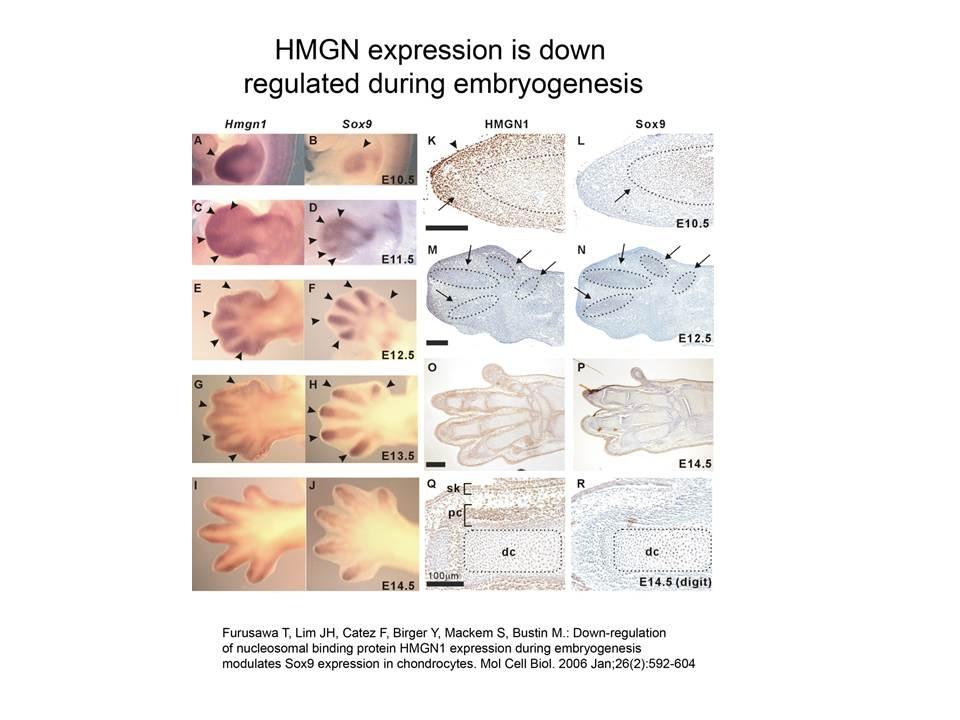
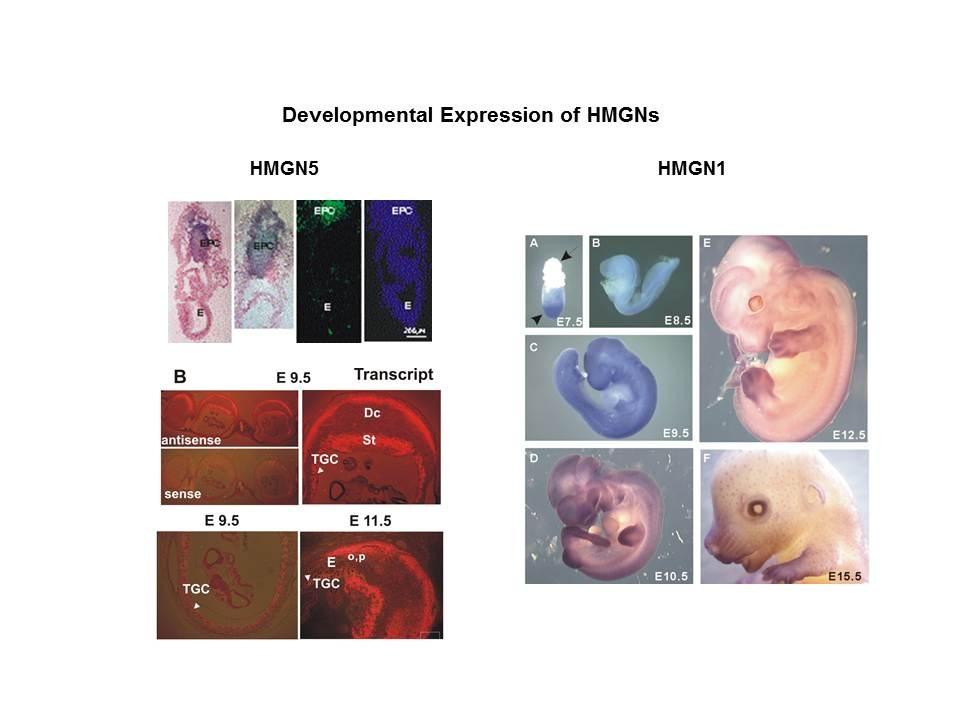
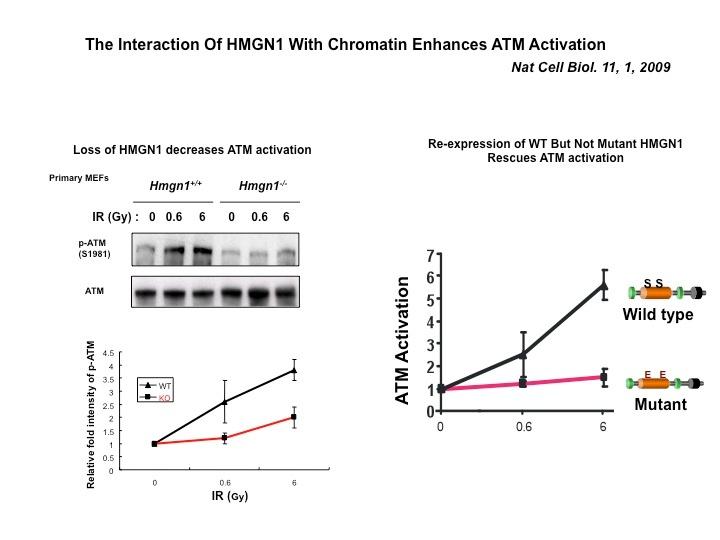
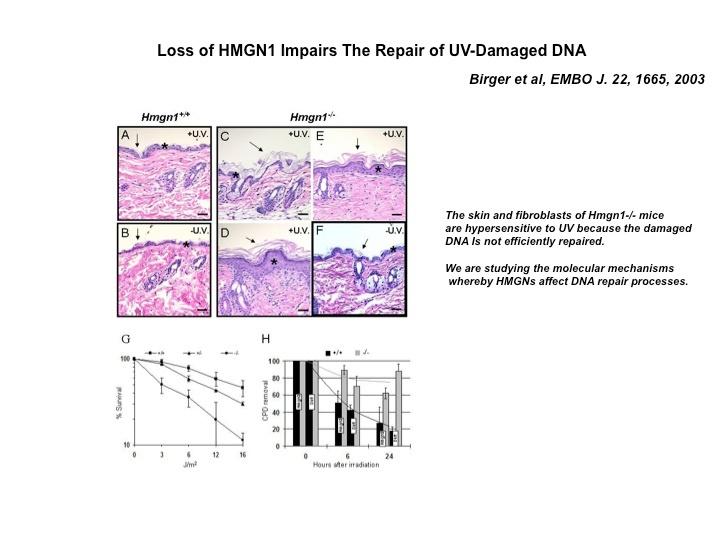
The Cellular Function of HMGN Proteins: Generation and Analysis of HMGN Knockout Mice. HMGN transgenic and knockout mice are used to investigate the biological function of HMGNs and the mechanisms whereby the interaction of HMGNs with chromatin impacts the cellular phenotype and play a role in development, differentiation, and disease.
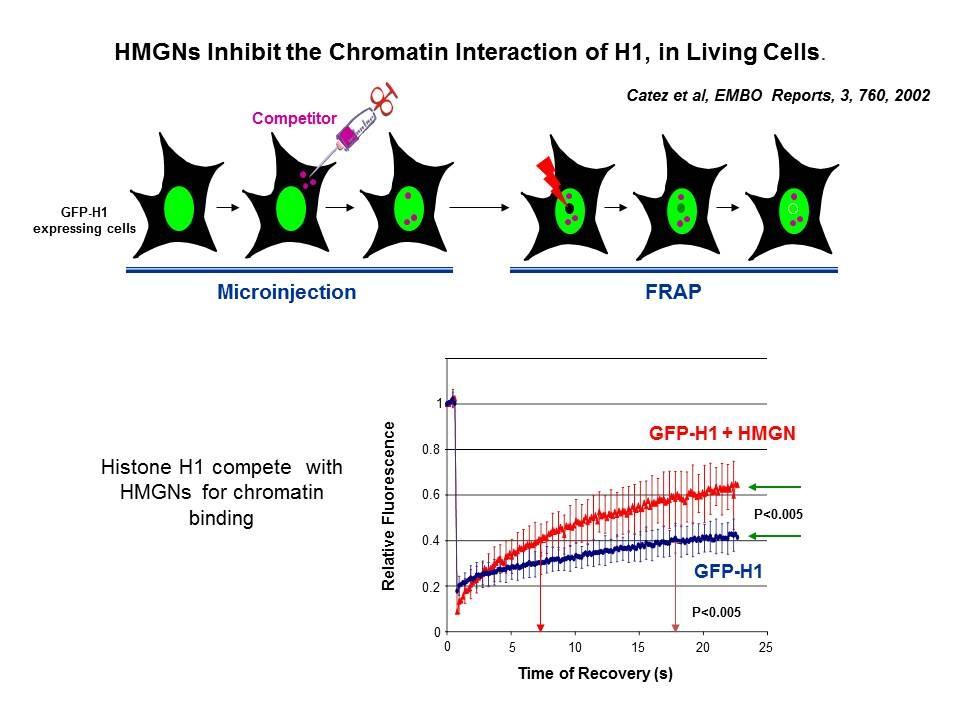
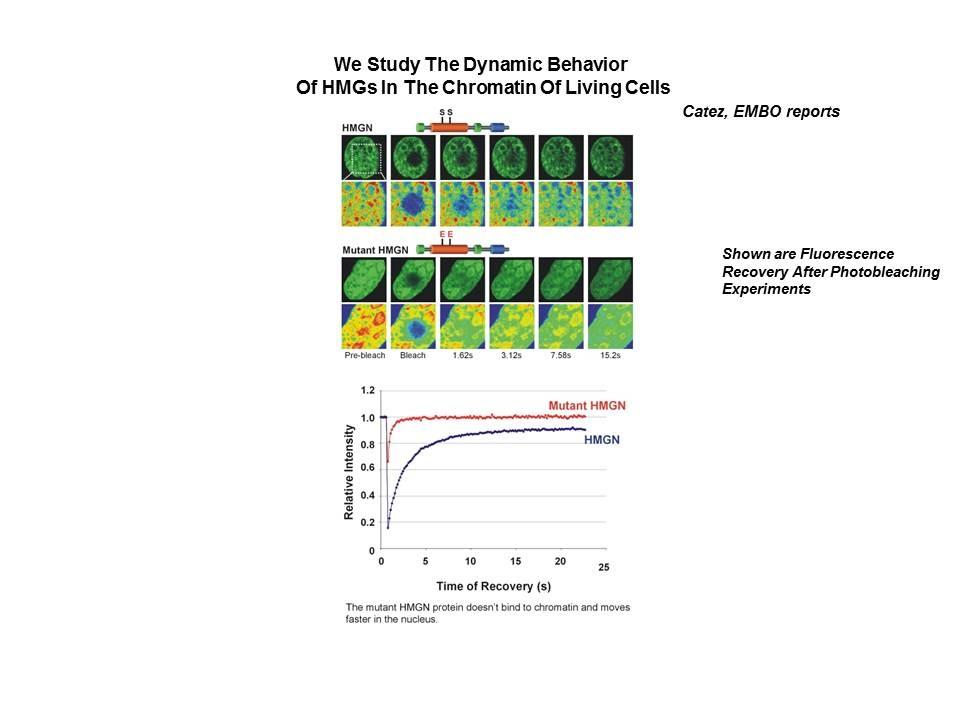
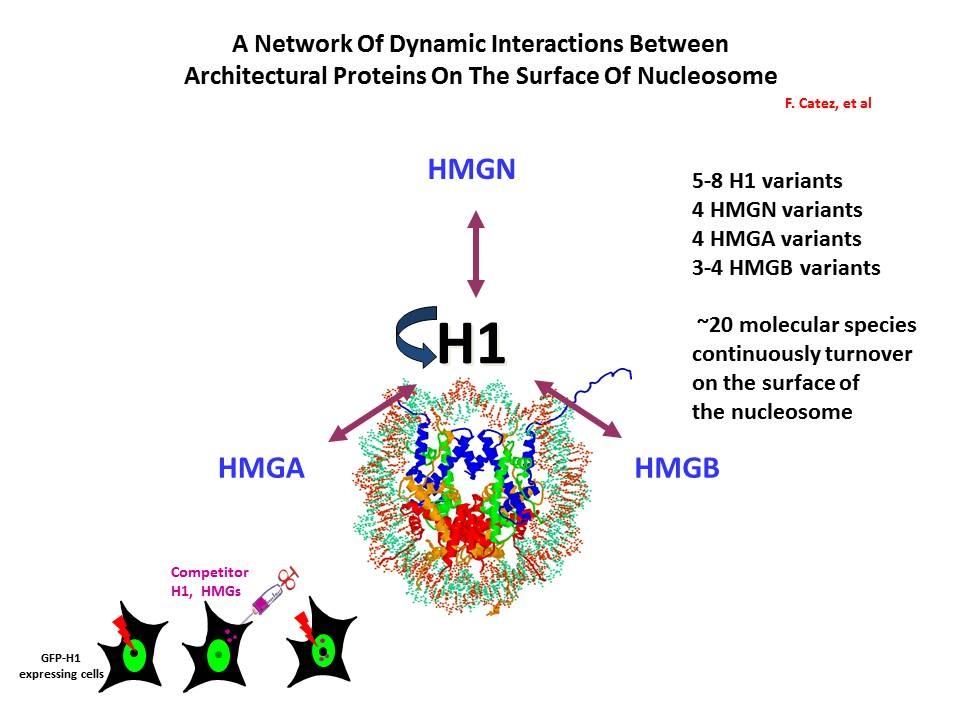
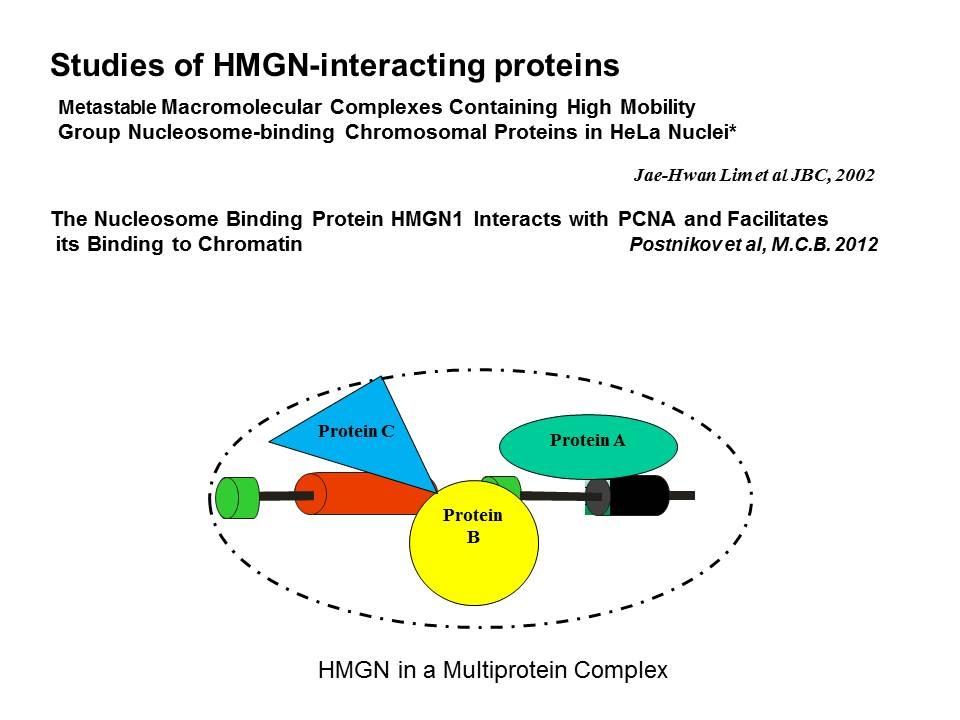
Linker Histone H1: HMG Interactions and Protein Networks in Living Cells. HMGNs inhibit the chromatin interactions of H1.
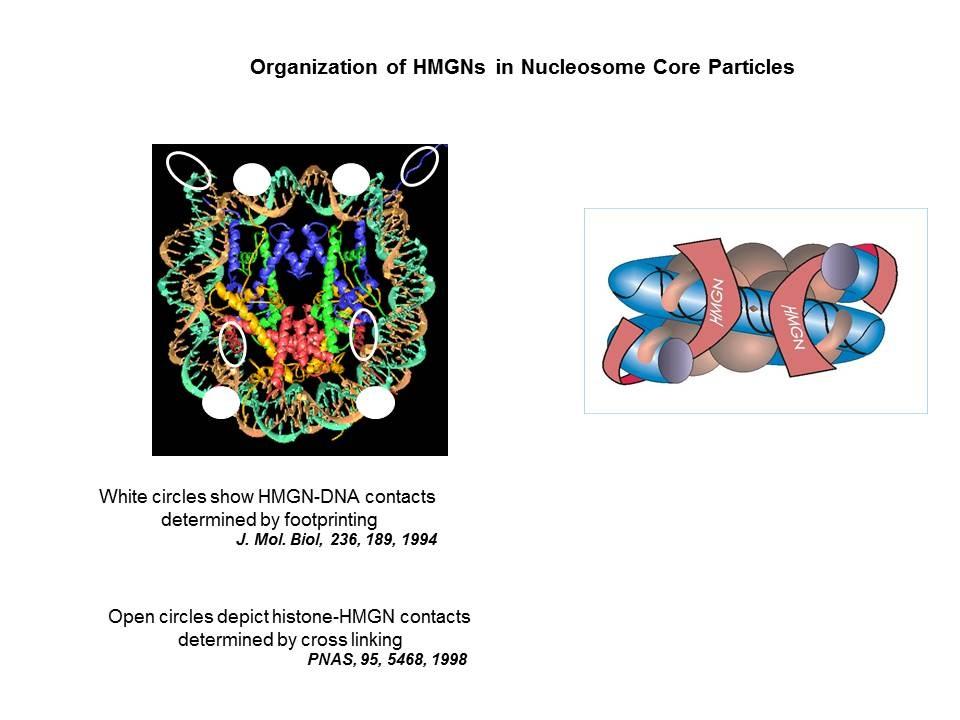
We demonstrated that the sites of interaction of HMGN with nucleosomes partially overlap the binding sites of histone H1 in chromatin (Alfonso PG, et al. J Mol Biol. 236:189-98, 1994).
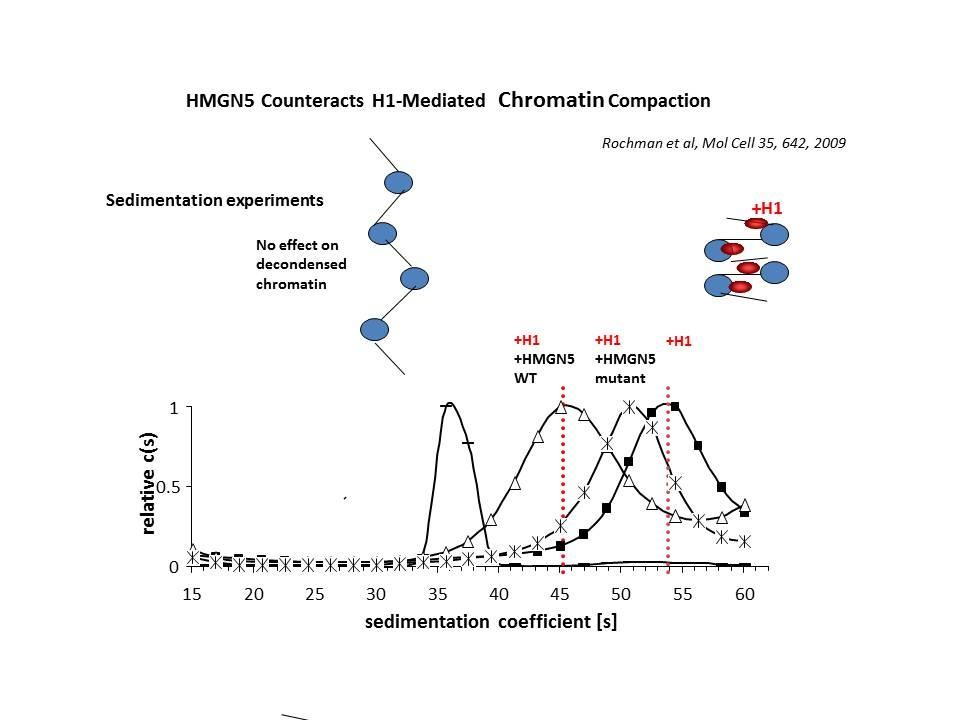
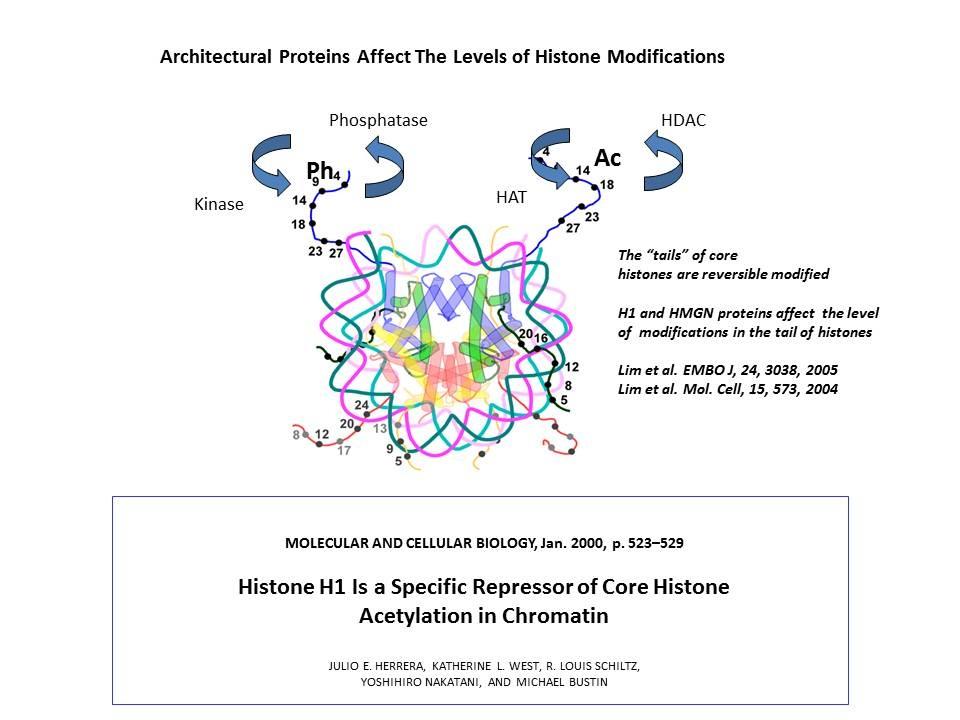
In collaboration with the Hansen Laboratory, we demonstrated that HMGN1 counteracts the repressive activity of histone H1 (Ding HF, et al. Mol Cell Biol. 17:5843-55, 1997).
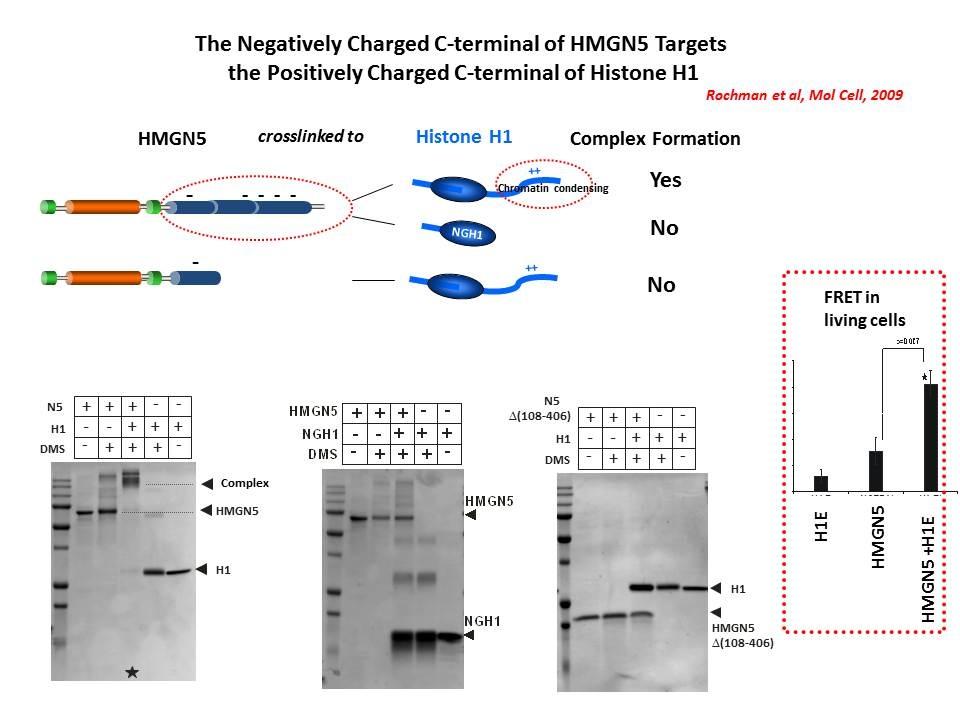
We demonstrated that HMGN5 competes with H1 and alters transcription and chromatin structure (Rochman M, et al. Mol Cell. 35:642-56, 2009).
We are interested in the mechanism whereby possible H1-HMGN interactions affect the structure and function of chromatin.
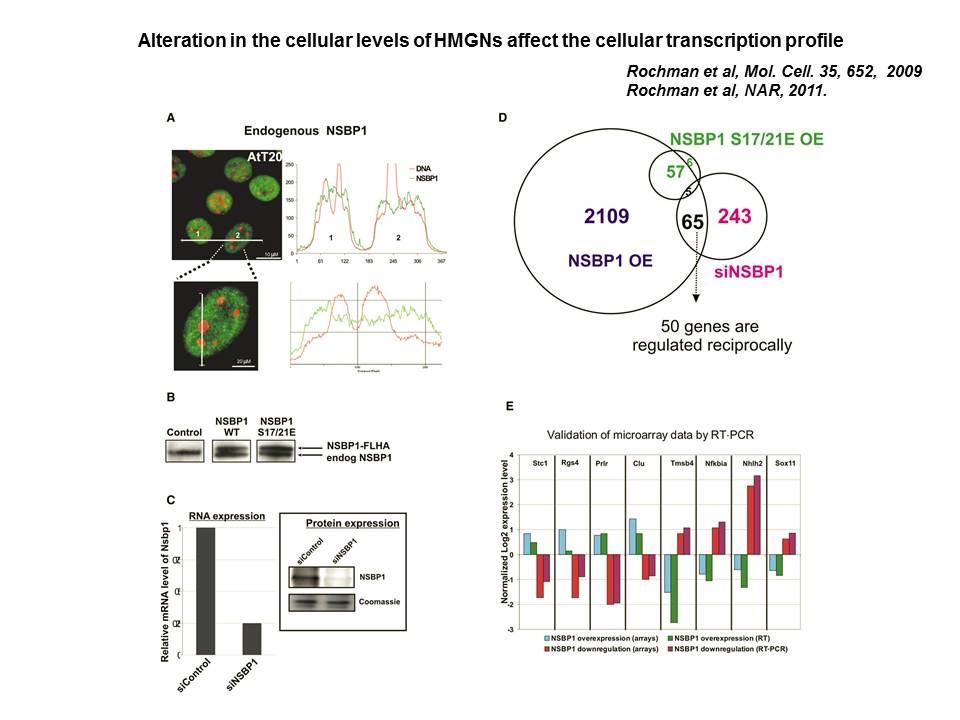
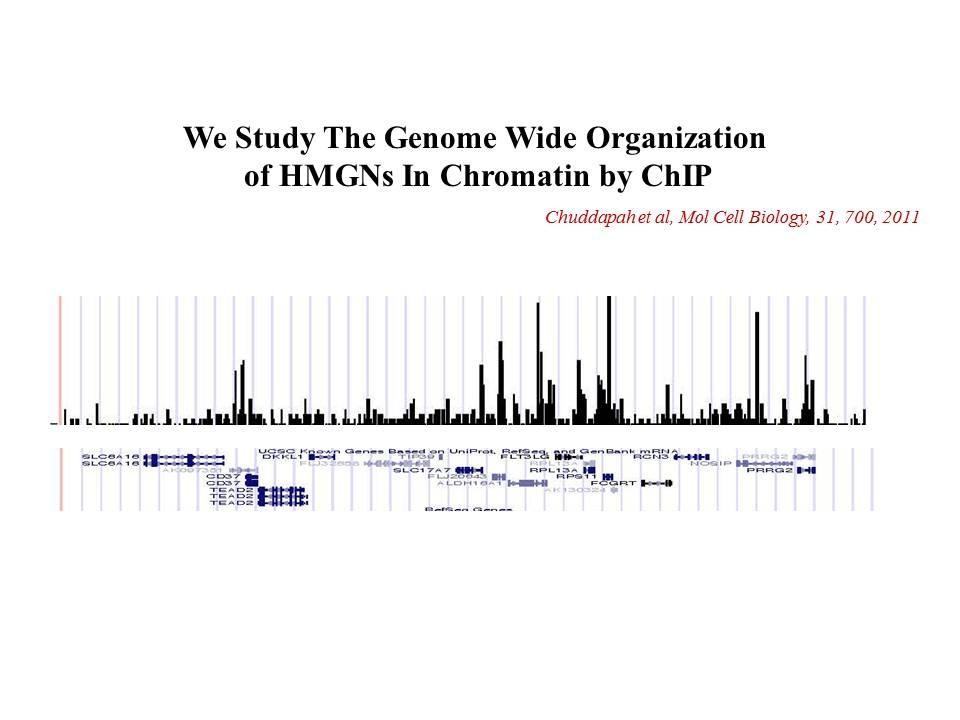
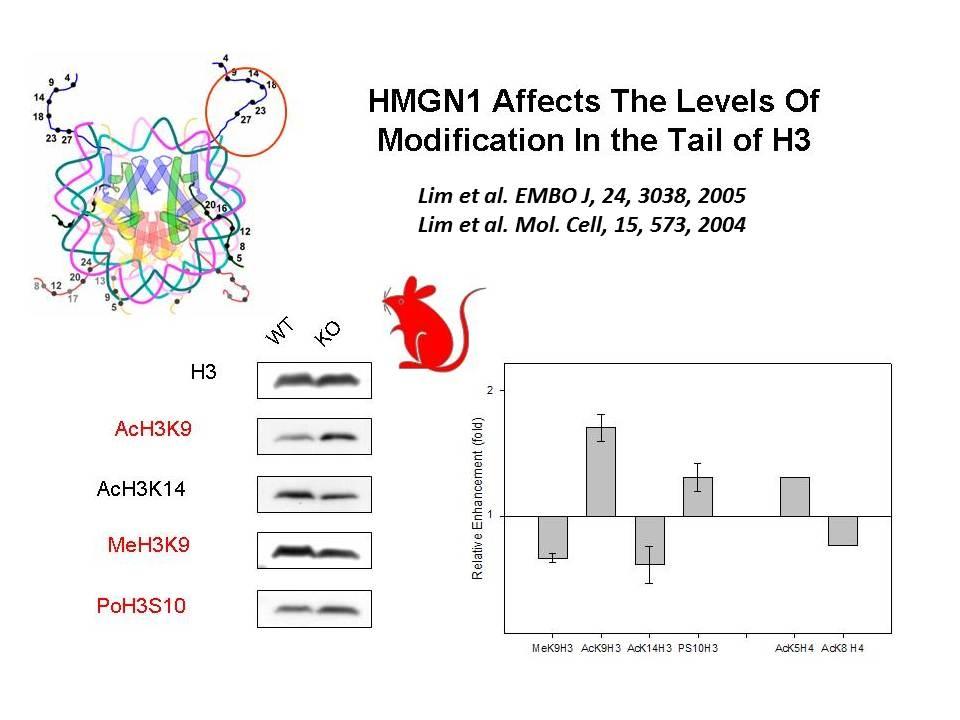
Role of Architectural Proteins in Epigenetic Regulation and Transcription. Alteration in the cellular levels of HMGNs affect the cellular transcription profile.
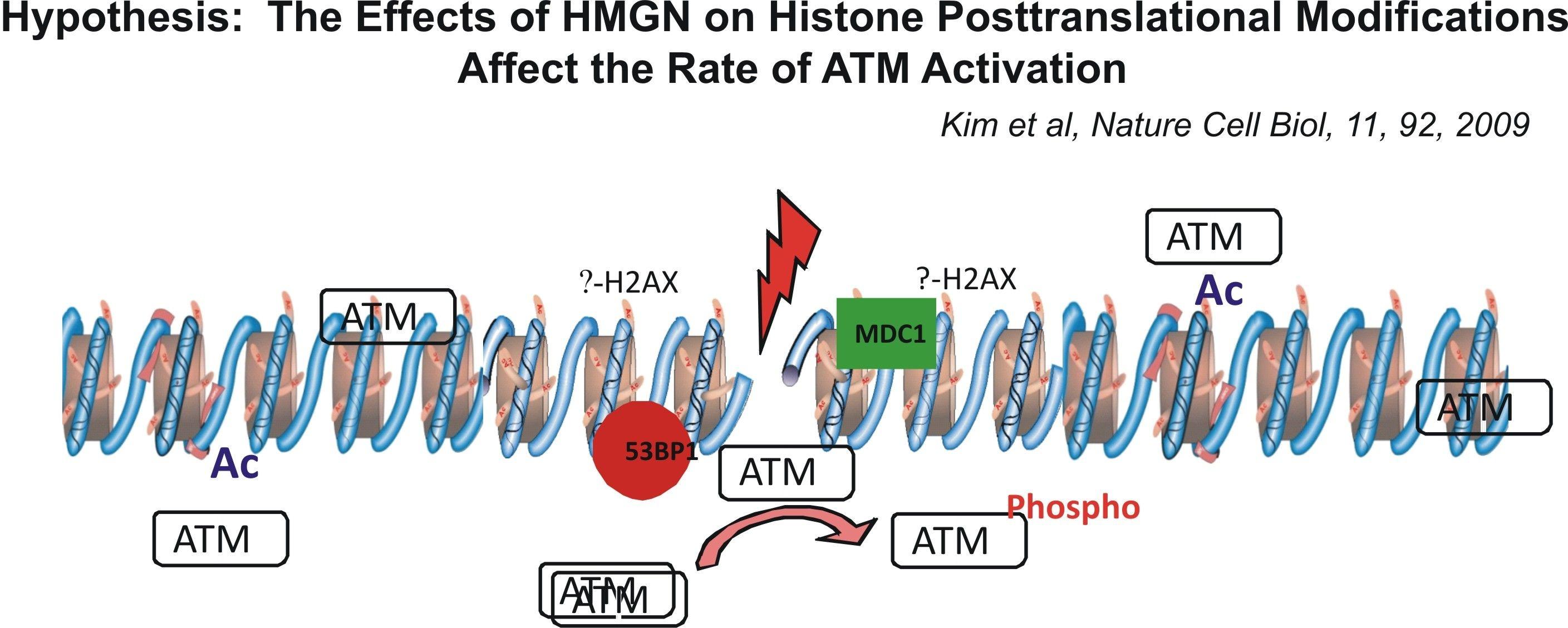
Using cells from knock-out mice and an in vitro reconstitution system, we find that H1 and HMGNs affect the levels of post-translational modifications in histone tails.