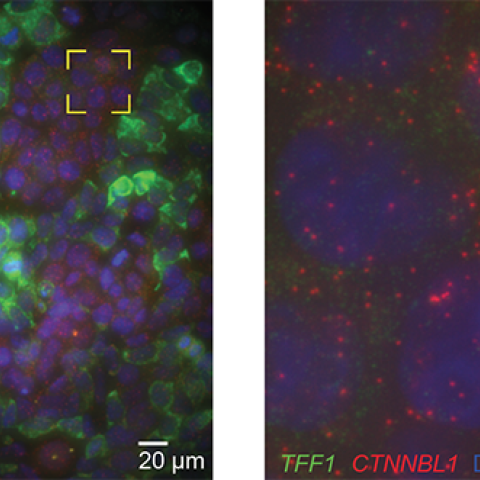
Periods of activity (left) and inactivity (right) of the TFF1 gene.
Photo courtesy of Daniel Larson
Researchers from CCR observed gene expression in real time and found, for the first time, that gene-directed RNA production is highly variable due to very long stretches of inactivity. The advent of CRISPR-Cas9 and technology to visualize single molecules of RNA helped make the findings possible. The research results were published December 13, 2018, in Cell.
The research supports the emerging awareness about the dynamic nature of gene expression and the tremendous variability among genes. A new observation that was particularly surprising was the long intervals between RNA activity and inactivity.
“We found that genes can turn off for hours or days at a time,” said Daniel Larson, Ph.D., Investigator in the Laboratory of Receptor Biology and Gene Expression. “This was a new degree of variation that we hadn’t been aware of before, and we don’t know why it happens.”
These findings were largely based on observations of the estrogen-responsive TFF1 gene in human breast cells. Larson and his team suspect that genes like TFF1, which code for secreted peptides, are more likely to have these long on-and-off lags. These genes also tend to be the noisiest, meaning that their behavior is particularly variable and random.
The research also showed an important relationship between signals such as estrogen and the structure of chromosomes found in the cell’s nucleus; chromosomal structure changes in response to estrogen stimulation. Using a model derived from theories about quantification, storage, and communication of information first developed about 70 years ago, Larson’s team developed a conceptual framework to show the degree to which cells can sense these signals.
The extent of gene variability has implications for understanding the functioning of cancer cells. “Heterogeneity is an important issue in cancer,” said Larson. “It can help explain why some tumors respond to immunotherapy and some don’t as well as the causes behind drug resistance. Being aware of intrinsic gene dynamics can also help identify when it is best to administer therapy.”
“There is no such thing as an average cell,” added Larson. “As sequencing becomes more sophisticated, it is increasingly important to understand the mechanisms of heterogeneity.”