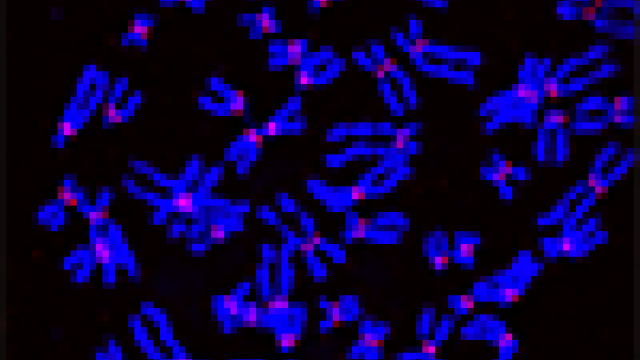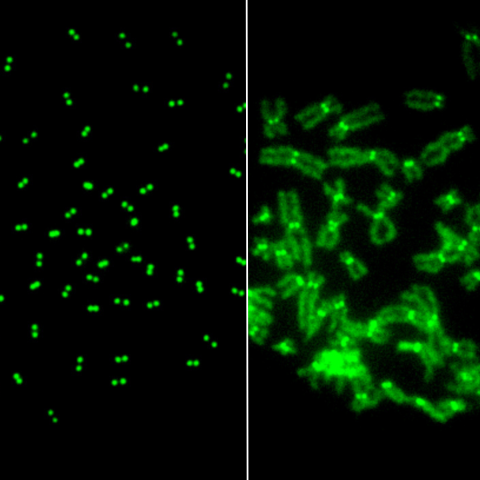
Mitotic chromosomes stained green for CENP-A in control (left) and DNAJC9-depleted cells (right), indicating higher CENP-A concentrations in DNAJC9-depleted cells.
Researchers have uncovered the causes and consequences of the overproduction and uneven distribution of CENP-A, a protein that is important for chromosomal stability. The results, published April 10, 2024, in the EMBO Journal, provide clarity on how excess CENP-A may play a role in cells that become cancerous.
In 2021, CCR researchers led by Munira A. Basrai, Ph.D., Senior Investigator in the Genetics Branch, discovered that overexpression of just one protein, CENP-A, contributes to uneven distribution of chromosomes during cell division and therefore leads to chromosomal instability. In healthy cells, CENP-A remains at the centromere, a region on the chromosome essential for the stability of the chromosome. However, overexpressed CENP-A can be improperly localized to other regions, a phenomenon referred to as mislocalization.
In this study, to find out the causes and results of CENP-A overexpression and mislocalization, the researchers performed a genome-wide screen looking for factors that contribute to the mislocalization of CENP-A. They found that CENP-A mislocalization was linked with the absence of a protein called DNAJC9.
DNAJC9 is directly involved with the correct folding of two histones, H3 and H4. These proteins are vital to maintaining the structure of chromatin and packaging the genetic material, explained Vinutha Balachandra, Ph.D., a postdoctoral fellow in Basrai’s lab. In this study, the researchers hypothesized that defects in the folding of histones H3 and H4 contributed to the mislocalization of CENP-A.
“Histones are building blocks, and not having histones formed properly is a bad thing for a cell,” said Basrai. “In this paper, we have shown that the histone defects seemingly allow CENP-A proteins the opportunity to localize where they shouldn’t be.”
Using a process called an interactome analysis, which is a study of how proteins work together, the team also showed that CENP-A hitchhikes a ride with another protein called MCM2, facilitating the mislocalization.
These findings are particularly significant because chromosomal instability is frequently found in many types of cancers. CENP-A overexpression is also correlated with poor prognosis, advanced disease stage, genomic instability and altered response to treatments.
“This study may lay the groundwork for future research into whether these mutations that are frequently observed in many cancers contribute to mislocalization of CENP-A,” said Basrai.
Currently, the Basrai group is working to learn more about the consequences of histone H3 mutations on chromosomal instability, as these mutations are found in several cancers. This work may help identify biomarkers that can aid in the prognosis of CENP-A overexpressing cancers.