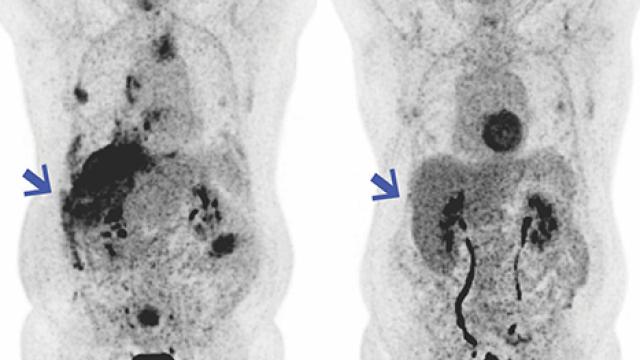
Mesothelioma is a rare but aggressive form of cancer which is difficult to treat. Image credit: iStock
Researchers at CCR have analyzed a rare type of cancer called mesothelioma in unprecedented detail, identifying gene signatures associated with low survival rates. The results, published February 10, 2023, in Cell Reports Medicine, also point to a subset of patients who respond well to certain therapies, which could help ensure more patients receive appropriate care moving forward.
“Mesothelioma is an aggressive cancer that can involve the lining of the lungs or the abdomen,” explains Raffit Hassan, M.D., a Senior Investigator in the Thoracic and GI Malignancies Branch. “Most patients have a poor prognosis, so we need to better identify those patients who respond to existing therapies and identify new therapies for treatment-resistant cases.”
To do so, Hassan’s team collaborated with Javed Khan, M.D., Senior Investigator in the Genetics Branch, and Eytan Ruppin, M.D.,Ph.D., Senior Investigator in the Cancer Data Science Laboratory, to analyze the genomic and transcriptomic nature of mesothelioma. At CCR, there is an established program for studying mesothelioma, so Hassan’s team was able to conduct a study and collect samples from a large group of 122 NIH patients. Notably, peritoneal mesothelioma, which affects the lining of the abdomen, is exceptionally rare and understudied compared to pleural mesothelioma, which affects the lining of the lungs. But in Hassan’s study, roughly half the samples were of peritoneal mesothelioma, allowing them to study differences between these two subsets of cancers.
Their data showed that – surprisingly – there are few genetic differences between pleural and peritoneal mesothelioma. Importantly, they identified a 48-gene signature in which high expression of these genes was associated with worse survival in both pleural and peritoneal mesothelioma. Higher expression of one gene in particular, CCNB1, was significantly associated with lower survival.
Next, the researchers wanted to analyze the data in order to find potential therapeutic targets for mesothelioma. Ruppin’s team had previously published a tool that could help with this analysis. The tool, SELECT, analyzes genetic data of tumors to find pathways that could be targeted to kill cancer cells, and cross-references that data with existing drugs that could potentially target those pathways.
SELECT revealed that one class of immune checkpoint inhibitors, called anti-PD1 therapy, and drug combinations involving the chemotherapy drug pemetrexed are effective therapies in two respective subsets of patients.
“We showed, as proof of concept, that we’re able to accurately predict which patient is going to respond to therapy, especially anti-PD1 therapy,” says Nishanth Nair, Ph.D., a staff scientist in the Cancer Data Science Laboratory who was also involved in the study. “This is very important because it could one day help ensure patients receive appropriate care in the clinic.”
Nair says he is excited at the potential for prospective studies to confirm the benefits of anti-PD1 therapy and pemetrexed in certain patients. The research team has made the data from their study freely available online, paving the way for more scientists around the world to study this rare form of cancer in greater detail.