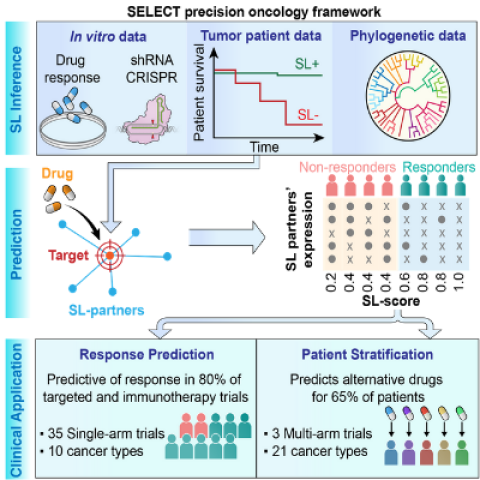
SELECT precision oncology framework.
Image Credit: Joo Sang Lee; Erina He, NIH Medical Arts
In an important advancement for precision oncology, CCR researchers have shown how their tumor analysis pipeline, called SELECT, can identify which therapies may be particularly beneficial for individual patients. When applied to data from over 30 different targeted and immunotherapy clinical trials, SELECT was predictive of patient responses to these therapies in about 80 percent of the trials. These findings were reported April 13, 2021, in Cell.
SELECT stands for SynthEtic LEthality and rescue-mediated precision onCology via the Transcriptome. The approach is based on identifying synthetic lethal interactions – a functional interaction between two genes whose concomitant inactivation kills tumor cells. These interactions provide an opportunity to selectively kill only tumor cells while sparing normal cells by targeting synthetic lethal pairs of specific genes inactivated in a tumor.
In the last 20 years, synthetic lethality has been investigated as a means to treat cancer, with some specific treatment regimens already used in the clinic. However, it is thought that many such treatment opportunities remain to be discovered, and SELECT offers a computational method for identifying such treatment options for individual patients. By analyzing tumor transcriptomics data, this approach can identify actionable tumor vulnerabilities that are not readily evident by traditional mutational- and gene fusion-based sequencing approaches.
Eytan Ruppin, M.D., Ph.D., Chief of the Cancer Data Science Laboratory, has been leading the development of computational tools to identify synthetic lethality. In recent years, he has used synthetic lethality to identify new drugs or gene combinations with therapeutic potential; a few of these studies have recently published, and one is already in clinical trials. In this most recent study, Ruppin, Kenneth Aldape, M.D., Chief of the Laboratory of Pathology, and colleagues took SELECT one step further by applying it to study the ability by which it can stratify cancer patients to treatments.
To test SELECT in this context, Joo Sang Lee, Ph.D., Sungkyunkwan University School of Medicine, South Korea, assembled a broad collection of 35 published transcriptomic datasets from targeted and immunotherapy clinical trials involving 10 different cancer types. He applied SELECT to predict the treatment response of the patients, given their tumor molecular data. He compared the patient recorded outcomes to SELECT’s predictions, finding them to be highly accurate in 80 percent of the trials.
“To the best of our knowledge, SELECT is the first approach that systematically achieves these moderate, but helpful levels of accuracy across many different therapies and cancer types,” Lee says.
“These results, while encouraging, need to be further studied in prospective studies where the predictions made will be used to inform clinicians’ treatment decisions,” Aldape says. This investigation is now underway in collaboration with several clinical teams at the NIH Clinical Center.
Ruppin hopes these prospective studies will further improve SELECT in the next few years, and if successful, establish SELECT as a complementary precision oncology approach for enhancing cancer-patient care.