Early Detection
Liver cancer patients suffer not only from the burden of tumors, but also from the compromised diseased state of the liver (chronic liver disease). The asymptomatic nature of early liver disease precludes our ability to effectively detect signs of liver cancer before significant disease progression.
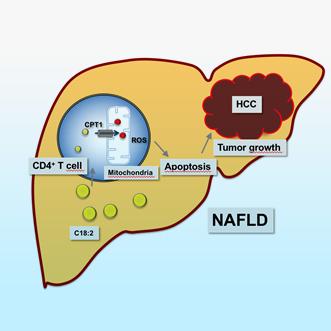
Therefore, strategies to identify liver disease status with proclivity towards tumor formation are in dire need. Current efforts include the detection and evaluation of novel biomarkers among patients with chronic liver disease, such as hepatitis viral infection, metabolic dysfunction-associate steatotic liver disease (MASLD/MASH; fatty liver), alcoholic liver disease.
Additional efforts aim to explore predictive signatures or algorithms for liver cancer initiation.
Please see related news and events and publications.
Diagnosis
Patients with liver cancer are currently diagnosed via imaging (ultrasound, CT scan and MRI), but most are at advanced stages of disease with limited treatment options. Liver function tests are also used in diagnosis, but parameters such as alpha-fetoprotein (AFP) are not sensitive or specific. Thus, novel diagnostic tools are highly warranted for liver cancer.
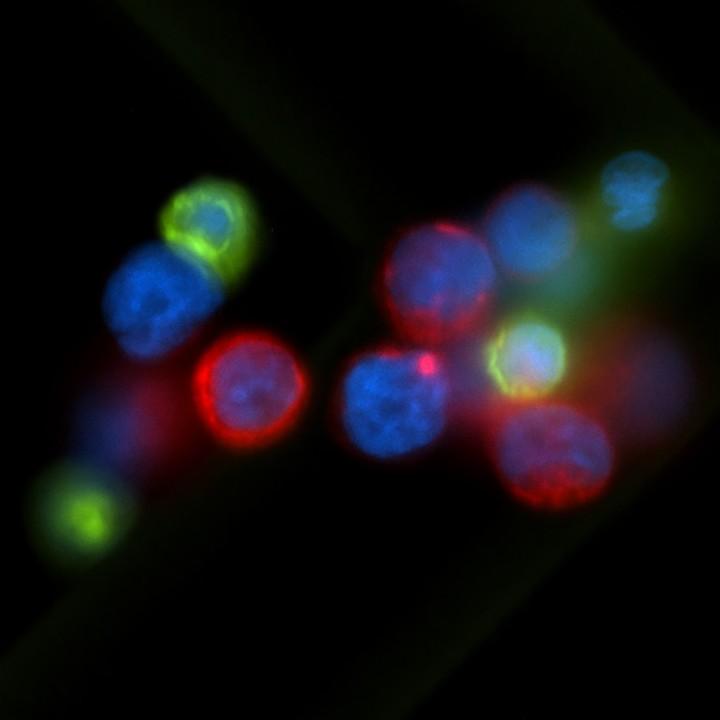
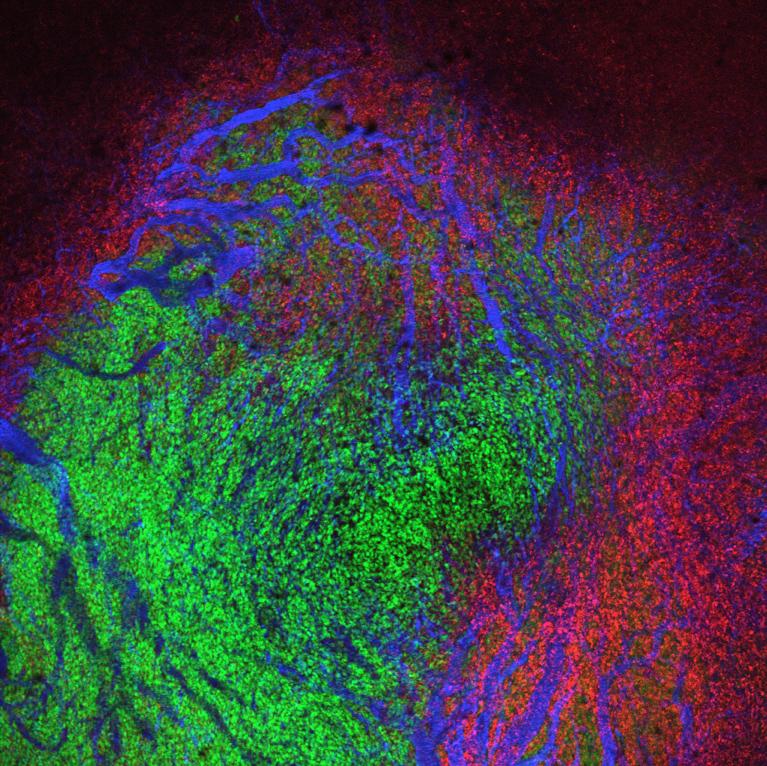
We are currently examining several parameters of tumor biology and the liver microenvironment, including cellular immunity and inflammation factors, metabolic species and the microbiome to identify biomarkers linked to liver cancer.
Approaches include, but are not limited to, exome sequencing of bulk tumors; single cell transcriptome sequencing of tumor core/resection biopsies and circulating tumor cells (CTC) as well as tumor-associated immune cells, tumor organoid culture models, PDX models, 3D-multiplex cell imaging-based technologies and artificial intelligence (AI)-guided multimodality imaging coupled to genomics (radiomics).
Please see related news and events and publications.
Prognosis
The current poor prognosis of liver cancer patients stems from multiple factors including its extensive heterogeneity, occurring between different patients, termed inter-tumor heterogeneity, and between different areas of an individual tumor, termed intra-tumor heterogeneity. Tumor heterogeneity may result from different cells of origin, etiology, underlying disease and diversity of genomic and epigenomic changes which drive tumor development. This results in varying degrees of clinical presentation and tumor biology, which impedes treatment options and poses a significant challenge to cancer management.
To address this problem, molecular-based technologies including genomic, transcriptomic, proteomic and metabolomic profiling, are being applied to identify distinct tumor subgroups with unique tumor biology which house critical and specific alterations in gatekeepers of cancer progression. Such molecular markers and signatures are being developed as prognostic tools to stratify patients for clinical management and predict outcome.
Please see related news and events and publications.
Treatment
One of main reasons why survival for patients with liver cancer has not improved in the past twenty years is the lack of approved novel therapies. Multiple phase III studies have failed to demonstrate any survival advantage for patients with liver cancer.
The advent of immunotherapy and biologicals and the implementation of precision cancer management strategy may dramatically shift this situation.
Our capacity to realign the immune response to improve patient response to treatment and outcome is being tested. Current pilot strategies include chimeric antigen receptors (CARs) and antibody conjugates such as Glypican-3 (GPC-3) and other means to enhance adaptive and innate immune responses in patients with HCC.
Please see related news and events and publications.
Population Studies
Liver cancer is a complex, multifactorial disease, contributing to a significant global health burden which varies among populations. The establishment of patient populations with well-annotated biobanks and well-characterized molecular features is essential to understanding its underlying biology, and developing effective strategies for its detection and treatment. To this end, we have successfully established large population-based studies including:
- The Thailand Initiative for Genomics and Expression Research in Liver Cancer (TIGER-LC) consortium aims to recruit 6,000 liver cancer patients in Thailand and matched high risk and population controls.
- The Maryland Liver Cancer Study aims to recruit 2,000 liver cancer patients in the greater Baltimore area in Maryland, U.S.A. at the University of Maryland at Baltimore and the Veterans Affairs (VA) Maryland Healthcare System, along with matched high risk and population controls (https://clinicaltrials.gov; NCT00913757).
These studies aim to identify genomic, genetic, etiological demographic, socioeconomic and environmental factors that modify liver cancer risk, susceptibility and progression. Comprehensive and comparative biological and clinical analyses will also promote a greater understanding of the environmental and etiological factors contributing to liver cancers across populations.
Please see related news and events and publications.
Collaborations
Expected Interactions and Synergies
The central idea of this proposal is to plant a seed, which allows for crystallization of different research activities in liver cancer research including scientists from NCI, NIH, academic institutions from the Washington area as well selected other extramural sites.
Engagement of Extramural Investigators
One of the central aims of the NCI-CCR-LCP is to foster collaborations between intramural and extramural investigators with an interest in liver cancer research. We plan to initiate and support these collaborations at every level.
NCI CLARITY Study
Cancers of the Liver: Accelerating Research of Immunotherapy by a TransdisciplinarY Network. The study aims to provide ‘clarity’ on who responds/does not respond to immunotherapy.
Contact us for additional information about our programs.