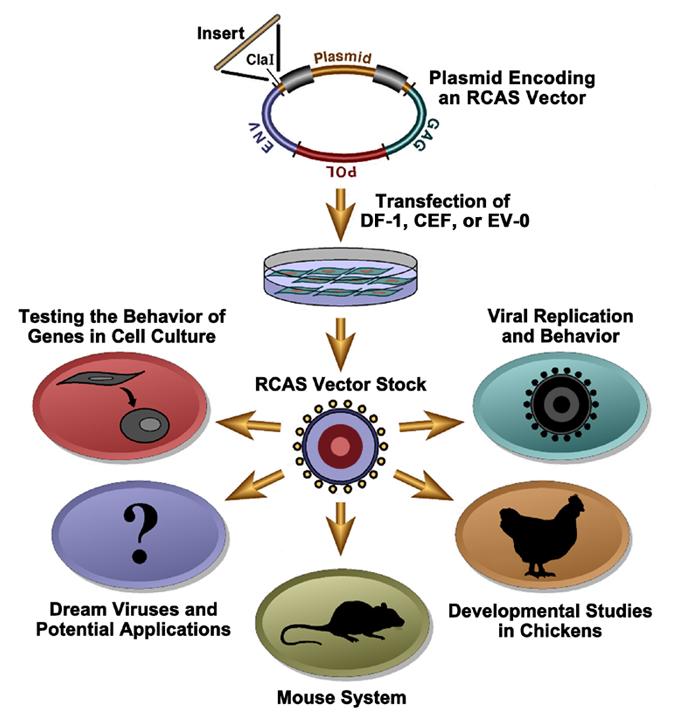
The RCAS vectors are a family of retroviral vectors derived from the SR-A strain of Rous sarcoma virus (RSV), a member of the avian sarcoma-leukosis virus (ASLV) family. In nature, retroviruses can acquire oncogenes from their hosts. In every case but one, acquisition of the viral oncogene has resulted in the loss of one or more viral genes. Most retroviruses that have acquired cellular oncogenes are replication defective; RSV is the exception. RSV contains a full complement of viral genes and the oncogene src. We have taken advantage of this exception to create a family of replication-competent retroviral vectors (see Table 1 and Table 2). Although the actual construction is more complex, the basic idea is that the RCAS vectors make it possible to substitute other genes/sequences for src. The name RCAS stands for Replication-Competent ASLV long terminal repeat (LTR) with a Splice acceptor. The RCAS vectors retain the src splice site and express an inserted gene via a spliced message. These vectors will replicate in appropriately chosen avian cells (usually chicken or quail, although others can be used). In some cases, appropriately chosen RCAS vectors will infect, but will not replicate, in appropriately chosen mammalian cells (see Table 3).
Different Types of RCAS Vectors
To make the RCAS vector system more broadly useful, there are vectors available in which several aspects of the basic vector have been changed. Table 2 describes these vectors.
Envelope/host range.
Retroviral infection requires the specific interaction between the envelope glycoprotein on the surface of the virus and the cognate receptor on the surface of the cell. The ASLV family of viruses has five primary envelope types: A, B, C, D, E. These recognize three distinct cellular receptors: A, C, and B/D/E. A full discussion of the various ASLV envelopes and their receptors is beyond the scope of this site. For additional information, we recommend the references provided in Chapter 3: Viral Entry and Receptors of Retroviruses edited by John M. Coffin, Stephen H. Hughes, and Harold E. Varmus, 1997, Cold Spring Harbor Laboratory Press, which is available as an online publication through the National Library of Medicine website. If you are undecided about which envelope to choose, A is usually a good choice; this envelope is not toxic to the host cell and viruses with this envelope usually have the highest titer.
Remember: In order for the virus to be propagated, your cells must have the appropriate receptor and cannot be sequentially infected with two ASLVs expressing the same envelope. Standard cells, the DF-1 chicken line (ATCC #CRL-12203) and the QT6 quail line (ATCC #CRL-1708 ) can be infected with subgroup A viruses. If you want to use chicken cells from a local source, make sure they are compatible with the vector you choose and make sure they are not already infected with ASLV.
If A is generally useful, why use other envelopes? Using RCAS vectors with two different envelopes (A and B, for example) makes it a simple matter to introduce two genes into a single cell (Givol et al., 1994, 1995, 1998). We have also prepared versions of RCAS vectors that use envelope genes from murine retroviruses. Although mammalian cells do not have functional receptors for the standard ASLV envelopes on their surface, the amphotropic versions of the RCAS vectors can infect (but will not replicate in) most mammalian cells. Alternatively, mammalian cells can be modified to express functional receptors for the A and B/D/E envelopes (see the Mouse System).
Expression.
As has already been mentioned, the RCAS vectors produce a spliced message that will lead to the expression of an appropriately inserted gene. The src-derived splice acceptor site is retained in the RCAS vectors; this means that expression of the spliced message for the inserted gene is driven by the viral LTR. For those who want to use another means to drive expression, there is the parallel family of RCAN vectors (Replication-Competent, ASLV LTR, No splice acceptor). The RCAN vectors differ from the corresponding RCAS vectors only in that they lack the src splice acceptor. RCAN vectors can express an inserted gene from an appropriate internal promoter; internal promoters can retain their tissue specificity when embedded in an RCAN vector (Petropoulos et al., 1992). RCAN vectors will also express an inserted gene if it is appropriately linked to an internal ribosome entry site (IRES); alternatively, a splice acceptor can be inserted (for example, using the adaptor plasmid SACla12Nco, described in Table 4. If you would like to express two (small) genes from a single vector, we recommend you use an RCAS (splice) vector and an IRES. There is also a gene trap RCANBP vector (pGT-GFP), which can be used in either avian or mammalian cells. This vector has a green fluorescent protein (GFP) insert in the opposite orientation to the viral genes. GFP is expressed when the gene trap vector is appropriately inserted into a host gene (Zheng and Hughes, 1999; see also Figure 1 and Gene traps and shuttle vectors below).
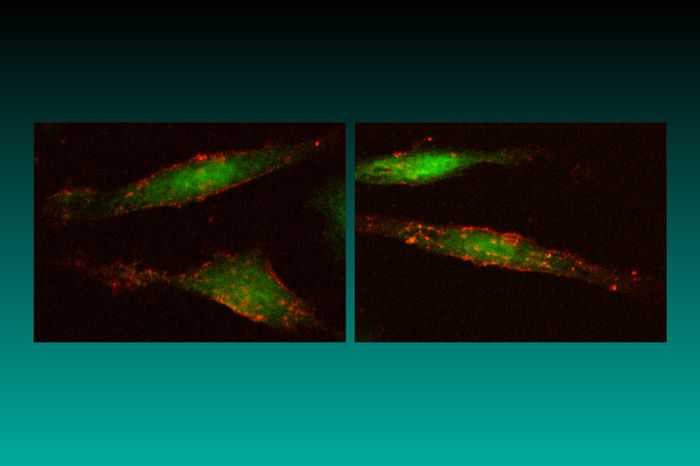
Figure 1. DF-1 cells expressing green fluorescent protein (GFP) and mouse heat-stable antigen (HSA). HSA is visualized by Texas Red immunofluorescence.
Replication efficiency/level of expression.
In some cases (for example, for some in vivo applications), it is helpful to be able to regulate the vector replication rate. It is also useful to be able to control the level of expression from the LTR, since this also determines (for the RCAS vectors) the level of expression of the inserted gene. There are two elements in the vector that contribute to the level of replication/expression. The first is the LTR. ASLV LTRs have strong enhancers; the promoter in the LTR is expressed at a high level in avian cells. Some mammalian cells express the ASLV LTR at high levels; others do not. We have some limited information on this issue; however, it is usually best to test the behavior of the ASLV LTR in a specific mammalian cell. The LTR from the corresponding endogenous avian retrovirus RAV-O appears to lack a strong enhancer; consequently, expression from the RAV-O LTR is much weaker than from the ASLV LTR. Vectors that contain the RAV-O LTR are RCOS and RCON (Replication-Competent, RAV-O Splice or No splice acceptor).
In addition to the LTR itself, the choice of the pol region also affects replication and expression in avian cells. (The region does not appear to have a strong effect on the level of expression in mammalian cells, however.) The original vector (RCAS) was derived in its entirety from the SR-A strain of RSV. Substituting the pol region from the Bryan high-titer strain of RSV produced a virus that replicated about one log better than RCAS in chicken cells. This derivative, now widely used, is called RCASBP (Bryan Polymerase).
Remember: RCAS and RCASBP are not the same vector. Please try not to confuse the two. Calling RCASBP RCAS is incorrect. In addition to RCAS and RCASBP, there is also RCOS and RCOSBP. In chicken cells, the difference between the level of each pair of these four vectors (in terms of replication and gene expression) is 5- to 10-fold. In order, from the lowest to the highest in terms of replication/expression, the vectors are RCOS, RCOSBP, RCAS, RCASBP. This gives the experimentalist about 4 orders of magnitude in terms of replication between the least efficient (RCOS) and the most efficient (RCASBP). The corresponding vectors that lack the splice acceptor follow the same order, from lowest to the highest in terms of replication/expression: RCON, RCONBP, RCAN, RCANBP.
Specialized RCAS derivatives.
Replication-defective derivatives. Although most RCAS vectors are replication competent, there are two replication-defective derivatives: BBAN and TFA-NEO. BBAN is based on a defective strain of RSV, the Bryan high-titer virus. Basically, BBAN contains a complete copy of Gag-Pol but is missing Env, so it can accommodate a larger insert in the ClaI site (~ 4 kb) relative to the conventional RCAS vectors (see below). It can be efficiently complemented by cotransfecting with an ASLV;env gene or with VSV-G. We do not have a complementing chicken cell line, and we no longer recommend the QT6 line originally developed for use with BBAN.
TFA-NEO is not, in the strict sense, a viral vector, but it is a transfection plasmid that is designed to accept (and express) the ClaI inserts prepared for use in the RCAS vector system. TFA-NEO was generated by removing the viral coding information (the segments between SacI and ClaI) from RCAS and inserting, in place of the coding region, a small oligonucleotide that contains sites for NsiI, EcoRV, and NdeI (in order from the SacI site). Warning: In TFA-NEO, the ClaI site is subject to dam methylation. To use the ClaI site, grow the plasmid on a dam- E. coli strain. To facilitate the selection of stable transformants in eukaryotic cells, TFA-NEO also expresses NeoR under the control of the chicken beta-actin promoter. The version of the promoter included in the plasmid is relatively weak; this favors the selection of cells that have the plasmid inserted in sites favorable for expression. The two expression cassettes (viral and beta-actin-neo) are separated by a polylinker (NsiI, SfiI, NotI, EagI) that makes it simple to generate a defined linear DNA for transfection of eukaryotic cells.
Gene traps and shuttle vectors. We have also prepared two types of specialized vectors: a GFP-based gene-trap vector (pGT-GFP) and two shuttle vectors (the RSVPs). pGT-GFP has a GFP gene inserted in the opposite orientation to the viral genes. The GFP coding region has a splice acceptor at its 5' end. Insertion of the pGT-GFP vector into the introns of expressed genes can generate a fused message that can lead to GFP expression (Zheng and Hughes, 1999).
Both of the RSVP shuttle vectors are based on RCASBP(A). One, RSVP(A)-Z, expresses zeocin resistance; the other, RSVP(A)-B, expresses blasticidin resistance. The RSVPs can be propagated either as viruses in avian cells or as plasmids in E. coli (the selections work in both systems). The RSVPs have, in addition to the selectable markers, a Lac operator sequence. This makes it easy to use the LacI protein to recover either integrated or unintegrated viral DNA, which greatly simplifies cloning. Details are provided in Oh et al., 2002.
Adaptors
The standard RCAS vector can be replicated in E. coli as a plasmid because it carries a pBR-derived replicon with an ampicillin-selectable marker. The entire plasmid is approximately 12 kb in length and has relatively few unique sites. All RCAS/RCAN vectors have a unique ClaI site for inserting a foreign gene. For those who are interested in the sequence, there is a second ClaI site, but it is methylated in Dam(+) strains of E. coli. There are RCAS vectors that have more than one unique site for inserting foreign genes; however, in most cases, genes/sequences are prepared for insertion into the ClaI site of RCAS vectors by using adaptor plasmids or PCR modification. Adaptor plasmids are small cloning vehicles that have multiple cloning sites flanked by ClaI sites. Your favorite gene can be converted into a ClaI segment by cloning it into an adaptor plasmid. The simplest adaptor plasmids are nothing more than a multiple cloning site flanked by ClaI sites and designed not to interfere with transcription or translation in the context of an RCAS vector (for example, Cla12). Adaptors can also provide a translational start site (Cla12Nco) and, if desired, a splice acceptor (SACla12Nco). Table 4 describes these adaptors.
Related Expression Systems
There is an E. coli expression system related to the adaptor plasmid Cla12Nco, which has a unique NcoI site (CCATGG). The ATG of the NcoI site is an efficient translation start site designed to give high-level expression in the context of the RCAS vectors. pUC12N is a high-copy bacterial plasmid that also has a unique NcoI site. The ATG in pUC12N is an efficient translation start site in E. coli. The polylinkers downstream of the NcoI sites are matched in Cla12Nco and pUC12N. Any insert set up for expression in one of these plasmids is automatically set up for the other system. The pUC12N expression system can be used to show that a particular insert is properly set up for expression in Cla12Nco; pUC12N can also be used to prepare material for immunization or biochemical analyses.
Insert Size
There are limits to the size of the insert that the RCAS vectors can carry. The limit is not defined entirely by insert size; some large inserts are tolerated much better than others that are the same length. There are few, if any, size-related problems with inserts smaller than 2 kb. Most, but not all, inserts up to 2.5 kb are well tolerated; most inserts larger than 2.5 kb are not. We have never received definitive reports of inserts larger than 3 kb that could be reliably replicated in an RCAS vector. If the insert is too large, part or all of the insert will be lost in the initial rounds of viral replication. A viral stock will be obtained (often with a brief delay); however, the recovered viruses will not contain the intact insert.
What to Avoid in Vector Design
- Don't include a transcription stop site or a polyadenylation site in your insert. It will interfere with viral replication; attempts to grow vectors with inserts that contain such sequences will lead to deletions in the insert.
- Avoid direct repeats in the inserted gene and make sure the insert does not have significant homology with the vector backbone. The mechanics of reverse transcription will cause deletions between directly repeated sequences anywhere in the viral genome (or insert). This process is efficient; the obvious way to avoid the problem is to avoid repeated sequences. The parental RSV has direct repeats flanking src in the RCAS vectors. The upstream copy of the repeat has been removed.
- Use inserts smaller than 2.5 kb (preferably smaller than 2 kb). Limits to the insert size are discussed above. Pushing the limits of insert size is usually more trouble than it is worth.
- Unless you want to create a cDNA, avoid splice sites in your inserts. The nature of the retroviral life cycle (alternating RNA and DNA genomes) gives the opportunity for splicing. While this can be helpful in generating cDNAs from genomic clones, it can also have unintended consequences. Remember: Some prokaryotic genes contain sequences that look like splice acceptor/donor sites to eukaryotic splicing machinery. Check the sequence carefully before you use it.
- It is usually quite difficult to propagate RCAS vectors that express inserts that are toxic to the host cell. If expression of the insert is harmful to the host, variants are rapidly selected that have deleted the insert. If you want to know if there is a problem with toxicity, put the insert in both an RCAS vector and the corresponding RCAN vector. If the insert is rapidly lost from the RCAS vector but not the RCAN vector, there is a problem with toxicity.
- RCAS/RCAN vectors work for expression of RNAi but not for antisense RNA (see Bromberg-White et al., 2004).
- If you use the vector in vivo, it can infect nondividing cells, but the titer is lower.
- Do not pass a viral stock from one cell culture to another unless it is unavoidable. With repeated passage, the insert will be lost. It is better to rederive fresh viral stocks by transfection.
- Keep in mind that the standard versions of RCAS are terminally redundant for the ends of the viral genome and contain two copies of the primer-binding site (PBS). (See also the section Plasmid Encoding an RCAS Vector.)
A Little Advice from the People Who Built the Vectors
The RCAS vectors were built to serve the needs of the research community. This webpage was created to help anyone who is curious about the system learn what it will (and what it won't) do and to help anyone who wants to use the system get started. If you have questions or problems and can't figure out the answers from this webpage and the literature, feel free to contact us. But, before you call or send e-mail, please look at the information provided on this webpage (especially the FAQs and literature sections; this will save us both a lot of time. If you have questions about a specific vector, please make sure you have the name exactly right, as it was when we sent it to you. Over the years, we have built hundreds of different RCAS vectors, but we cannot answer any specific questions about the vector if we don't know what it is you have. If you did not receive the vector from us, we cannot help you. There are a considerable number of derivative RCAS vectors that we did not create. Some of them are useful; others are not. We have no way of knowing which are which.

An RCAS plasmid is shown in the diagram above. The viral sequences are arranged in the form of a provirus; the bacterial plasmid sequences (which are derived from pBR322) lie between the long terminal repeats (LTRs). The plasmid confers resistance to ampicillin; details of the construction are given in Hughes et al., 1987. We have provisional sequences (downloadable as text files) for several of the commonly used RCAS vectors: RCASBP(A), RCASBP(B), and RCASBP(M). Numbering of these sequences begins at the U3 R boundary in the lefthand LTR. Note that the sequences have not been finalized. We are in the process of preparing the final versions of the sequences. As soon as this has been done, the corrected sequences will be posted here.
The RCAS family of plasmids were originally derived from a molecular clone of the SR-A strain of Rous sarcoma virus. The molecular clone was a two-LTR circle. When this circular viral DNA was converted into proviral form, a small amount of viral sequence was retained adjacent to each LTR. Approximately the first 130 bases upstream of the left-hand LTR and 150 bases downstream of the right-hand LTR are of viral origin. For those who use the vectors as gene delivery vehicles, these viral sequences are usually irrelevant. However, for those who use the vector to make and study the effects of mutations on viral replication, these duplicated viral sequences can be important. For example, the viral sequences downstream of the right-hand LTR contain an intact primer-binding site (PBS) and the viral sequences upstream of the left-hand LTR contain a second copy of the PBS. Anyone wanting to study the effects of mutating these sequences should be aware that the second copy of these sequences can confound the analyses. We have prepared a version of RCASBP(A) in which these redundancies have been removed. This vector, RCASBP(A) DeltaR, is available upon request.
Table 1. Standard RCAS Vectors
Table 2. Other RCAS Vectors
Table 3. RCAS Vectors for Use in Mammalian Cells
Table 4. Adaptor Plasmids

To avoid recombination with any of the closely related endogenous retroviruses commonly found in chickens, we recommend the use of cells derived from the EV-0 strain of white leghorns. This strain is maintained at the Regional Poultry Research Laboratory (RPRL) in East Lansing, Michigan (telephone 517-337-6828; fax 517-337-6776). Fertilized eggs can be obtained from the RPRL for a nominal charge. In many cases, it is convenient to use the permanent EV-0-derived chicken cell line DF-1. DF-1 cells were prepared by Doug Foster (University of Minnesota) and can be obtained from the American Type Culture Collection (ATCC; catalog #CRL-12203). Note that this cell line is not listed in the ATCC website, but is available through telephone orders (800-638-6597 or 703-365-2700).
We have tried a number of transfection protocols. Most will work; the standard calcium phosphate protocol is efficient and reliable. Start with 5-10 micrograms of purified RCAS plasmid DNA. Originally, we used cesium chloride-banded DNA; for most purposes, we now purify the DNA by using Qiagen columns. (A detailed transfection protocol will be provided soon at this webpage.)
The virus is unstable; treat it gently. Freshly harvested virus can be stored for a day or two at 4°C; for longer periods, freeze at -70°C. We usually prepare fresh virus by transfection rather than by passaging a virus stock. All retroviruses are genetically unstable; therefore, if the viral stock is passaged, the inserts will be lost. This is not (usually) a problem if the vector carries a gene for a nontoxic protein and the vector stock has been freshly prepared by transfection.
In some cases, it is convenient to use cocultivation to transfer virus from one cell to another. This is a particularly useful and efficient approach if the target cell is a mammalian cell in which the RCAS viruses will not replicate. The avian cells that were used to produce the virus can be removed from the culture if the target (mammalian) cells have a selectable marker or a cell surface antigen that can be used to distinguish them from the avian cells. Alternatively, the avian producer cells can be treated with mitomycin C. This blocks further cell division, but the treated cells continue to produce virus.

Ordinarily, a vector stock is obtained simply by pipetting off the medium from an infected culture. The titer of the virus in the culture medium is sufficient for most purposes. For those who want a more efficient infection, there are two simple alternatives: either the virus in the medium can be concentrated (usually by sedimentation in an ultracentrifuge), or infection can be carried out by cocultivating the target cells and the virus-producing cells. Each protocol has particular advantages and disadvantages.
Cocultivation is simple and effective; however, the producer cells usually need to be removed from the culture at some point. If the target cells express a selectable marker, the cells can be separated using the appropriate selection procedure. Alternatively, the virus-producing cells can be treated with mitomycin C before the target cells are added. Mitomycin C treatment “kills” the cells (they cannot be passaged), but they remain metabolically active and continue to produce virus with only a modest reduction in titer.
Concentrating the virus by sedimentation also works, but the recovery of infectious virus is not always complete. Quite often, the physical concentration is higher than the concentration of the viral titer, probably due primarily to the fact that the viral pellet is difficult to resuspend. Some people prefer to sediment the virus onto a dense sucrose cushion rather than the bottom of a tube. In any case, do not subject the virus to significantly more sedimentation than what is needed to pellet the virions. When resuspending the concentrated virus, be patient and gentle. Let the pellet sit on ice, then resuspend it by gentle pipetting (a pipetteman works well) rather than using prolonged or vigorous vortexing.

The available RCAS vectors allow one to introduce a variety of genes into any cultured mammalian or avian cell. There are some advantages to using the RCAS system to introduce genes into cultured cells, particularly into avian cells. Because the vector replicates, it spreads rapidly to essentially every cell in the dish. If an efficiently replicating vector (such as RCASBP) is used, this usually takes about 1 week. No selectable marker is used, and there are no problems related to picking out one cell or a few cells that may not share all of the properties of the cells in the original culture. Chicken embryo fibroblasts (CEFs) are easy to prepare; experiments with CEFs can be carried out in primary cells as well as in cell lines. If you are willing to work with a continuous cell line, DF-1 is relatively easy to handle and is available through the ATCC. The RCAS vectors are available with several distinct envelopes, so that it is possible to doubly (or even triply) infect a culture. This allows one to look at the interactions of two (or more) genes in cultured cells. Similar experiments can be done in vivo (Givol et al., 1994, 1995, 1998). We have also developed RCAS vectors that use an IRES to express two proteins (see Figure 1 below).

Figure 1. DF-1 cells expressing green fluorescent protein (GFP) and mouse heat-stable antigen (HSA). HSA is visualized by Texas Red immunofluorescence.
There are a few obvious pitfalls in experiments of this type. If you are trying to overexpress a gene that will strongly interfere with the replication of the host cell (or the viral vector), you are likely to have problems. Basically, the expression of a gene that strongly interferes with the replication of the host cell sets up a strong selection for the viral vector. Among the variant viruses that arise, any that do not express the gene of interest will replicate much more rapidly than viruses that express the gene. This gives rise, usually quite rapidly, to virus stocks that fail to express the gene. We have been able to do some experiments with RCAS vectors expressing genes that block cell division (Givol et al., 1995, 1998), but the experiments — and the cell culture — have been much more difficult than usual.
Over the years, a number of laboratories have tried to use the RCAS system to block the expression of host cell genes by providing antisense transcripts. So far as we know, all of these attempts failed; therefore, we do not recommend using the RCAS vectors for expressing antisense transcripts. However, the vectors can be used to express siRNAs (Bromberg-White et al., J. Virol. 78: 4914-4916, 2004).

The RCAS vectors are simple, replication-competent avian retroviruses. They adhere to the rules that govern retroviral replication. For this reason, the RCAS vectors are widely used as tools to study retroviral replication. Including a scorable (GFP, AP, etc.) or a selectable (puro, neo, etc.) marker in the vector makes it much simpler to monitor the viral titer and viral replication. The retroviral life cycle is shown schematically in Figure 1 below. The life cycle begins with the binding of the envelope glycoprotein on the surface of the virus to a cognate receptor on the surface of the target cell. This interaction leads to the fusion of the viral and cellular membranes. Although in the figure this process is depicted as occurring at the plasma membrane, recent data suggest that the ASLVs may use a pathway similar to that used by influenza virus and enter the cell through a low-pH mechanism (Mothes et al., 2000).
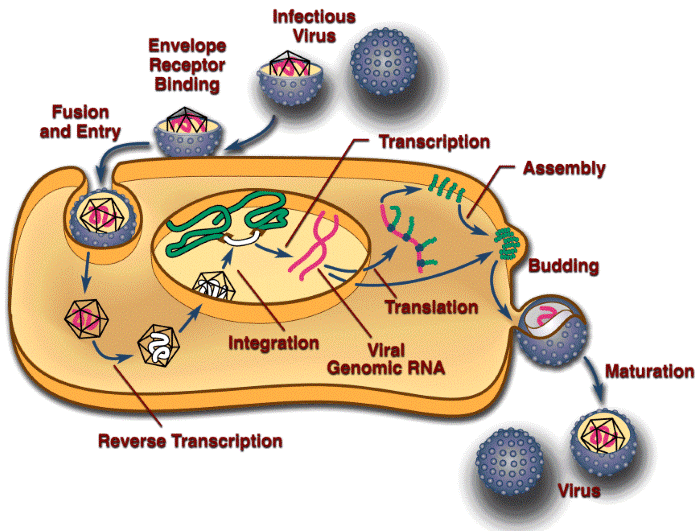
Figure 1. A diagrammatic representation of the retroviral life cycle.
After viral and cellular membranes fuse, the viral core is in the host cytoplasm, where the single-stranded RNA genome of the virus is converted into double-stranded linear DNA by the enzyme reverse transcriptase. Understanding the mechanics of the reverse transcription process is important for good vector design (see Overview – What to Avoid in Vector Design). However, the details of the reverse transcription process are beyond the scope of this website. If you would like to learn more about reverse transcription, please refer to Retroviruses (edited by John M. Coffin, Stephen H. Hughes, and Harold E. Varmus, 1997, Cold Spring Harbor Laboratory Press), which is now available as an online publication through the National Library of Medicine website.
Once the double-strand viral DNA is synthesized, it must move from the cytoplasm to the nucleus. The viral DNA of some complex retroviruses, such as HIV-1, can transit the nuclear membrane. The viral DNA of some simple retroviruses, including MLV, does not transit the nuclear membrane and these viruses cannot infect nondividing cells. However, it was recently shown that ASLVs (including RCAS vectors) can infect nondividing cells in culture, albeit at a reduced efficiency relative to infection of dividing cells (Hatziioannou and Goff, 2001; Katz et al., 2002).
Once the viral DNA has access to the host genome, integration can take place. Integration links the ends of the linear viral DNA to host DNA. The reaction is specific with respect to the viral sequences involved and nonspecific with respect to the host sequences. The site of integration in the host genome influences the expression of the integrated provirus. Although the provirus has its own promoter, the activity of the viral promoter is influenced by the adjacent host sequences. Conversely, the insertion of the provirus can also influence the expression of host genes. The provirus is an insertional mutagen: it can disrupt a gene by inserting into it, or can either enhance or decrease the expression of gene by inserting next to it. Because higher eukaryotes are diploid, this ordinarily does not matter; however, in some cases, insertion near a cellular oncogene can lead to oncogenesis in infected animals. Because the viral DNA becomes part of the host genome, infection is permanent. The infected cells, and all their progeny, will carry the inserted viral sequence. If the infection takes place in a germ cell, or a germ cell precursor, the resulting animal will carry the virus as a transgene (usually called an endogenous provirus).
It is sometimes useful, in studying viral replication, to recover either unintegrated or integrated viral DNA. With conventional retroviruses (or retroviral vectors), recovery usually involves PCR amplification and/or cloning. However, it is possible to generate retroviral shuttle vectors that can be propagated either as viral stocks or as plasmids in E. coli. This makes it easy to recover either unintegrated or integrated viral DNA. We have prepared a set of RCASBP(A)-derived shuttle vectors, the RSVPs. These vectors are listed in Table 2; a more complete description is provided in Oh et al., 2002.
Once embedded in the host genome, the provirus behaves like a cellular gene. For simple retroviruses, such as ASLVs, expression is entirely under the control of the host’s machinery. The RNA is transcribed by the host’s DNA-dependent RNA polymerase. Unless the experimentalist has inserted an internal promoter, there is, for simple retroviruses, one primary transcript that begins at the U3 R junction in the upstream LTR and terminates at the R U5 junction in the downstream LTR. This full-length RNA is exported from the nucleus and serves as genomic RNA and as the message for the Gag and Gag-Pol polyproteins. A portion of the full-length RNA is spliced to produce the message for the Env protein, and in the case of RSV and the RCAS vectors, there is a second spliced message (see Figure 2B below), which leads to the expression of the src gene in RSV or the inserted gene in RCAS.
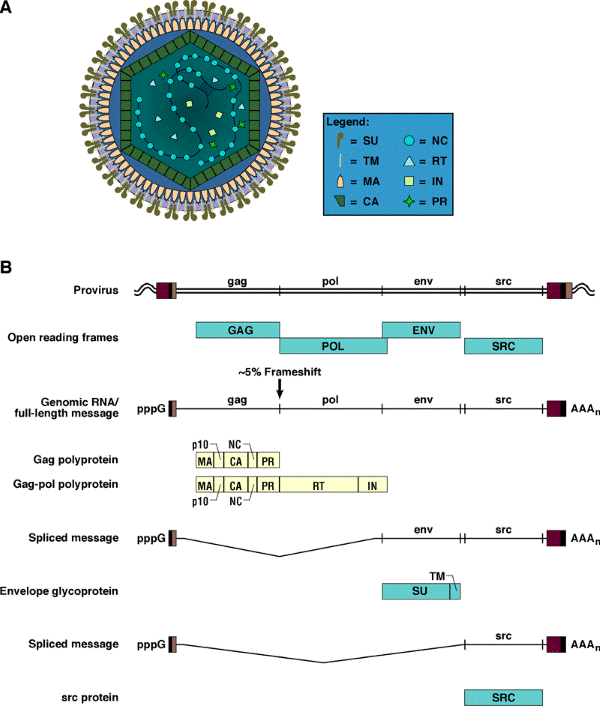
Figure 2. RSV virion structure and expression. Panel A shows a diagrammatic representation of a mature Rous sarcoma virus (RSV) virion. The legend at the right identifies individual proteins found in the mature virion. The outer membrane of the virion contains the transmembrane protein (TM), which is associated with the surface protein (SU). The matrix protein (MA) lies just under this outer membrane. The core of the virion is structurally delimited by the capsid protein (CA). Inside the capsid are two viral RNA genomes, shown partially covered with nucleocapsid protein (NC). The two genomic RNAs are hydrogen bonded near their 5' ends. The core also contains reverse transcriptase (RT), integrase (IN), and protease (PR). Panel B shows the relationship of the proviral DNA, the open reading frames, viral RNAs, and proteins of RSV. The LTRs of the provirus are shown as a series of three boxes (U3, R, and U5). The viral genome is divided into gag, pol, env, and src genes. Adjacent host DNA is shown as a wavy double line. In the case of RSV, the gag and env genes are in the same reading frame, pol and src are in a different reading frame. Host DNA-dependent RNA polymerase transcribes the provirus, yielding a full-length RNA that serves both as the viral genome and as the message for the gag and gag-pol polyproteins. This RNA is capped (indicated by pppG) and polyadenylated (AAAn). Approximately 5% of the time full-length RSV RNA is translated, there is a frameshift event that allows the ribosome to synthesize the gag-pol polyprotein. The gag and gag-pol polyproteins are processed by the viral PR to yield the proteins found in the mature virion. The relative positions of these components in the polyproteins are shown. The RSV polyproteins contain a p10 protein whose function is not well understood. A portion of the full-length viral RNA is processed by the host splicing machinery, which gives rise to the env and src mRNAs. The env mRNA is translated into the envelope glycoprotein, which is cleaved into SU and TM by a host cell protease. The src mRNA is translated into the src oncoprotein.
The messages are translated in the cytoplasm. The gag gene is the 5'-most gene in unspliced RNA. In the case of the ASLVs, the pol gene is in a different reading frame. During ~5% of Gag translation, a frameshift suppression event causes a ribosome near the gag-pol boundary to slip over into the pol gene. This leads to the synthesis of the Gag-Pol polyprotein. Gag and Gag-Pol self-assemble at the plasma membrane, picking up two copies of viral genomic RNA and some small host RNAs — in particular, the tRNA trp, which is used to initiate reverse transcription. The assembled virus buds through the plasma membrane of the host cell. In so doing, it not only acquires a membrane, but picks up the envelope glycoprotein along with it. Either during or shortly after budding, the viral protease cleaves the Gag and Gag-Pol polyproteins, giving rise to infectious virus. The newly budded virus cannot (usually) reinfect the cell that released it. Ordinarily, the infected cell produces enough of the envelope glycoprotein to block its own receptors, a process called receptor interference.

Relative to the mouse system, the chicken system has significant advantages, and substantial disadvantages. On the plus side, the developing chicken embryo is relatively large, and the fact that it develops outside the mother makes it much easier to manipulate. On the minus side, chicken genetics are considerably less powerful than mouse genetics. We have tried to make it possible to use the RCAS vectors both in the avian system and in the mouse system, so that the experimentalist can have the best of both worlds.
The experiments done with chickens can be (somewhat arbitrarily) divided into those done with embryos, and those done post-hatch. For the most part, experiments done post-hatch are intended to look at biological questions or to investigate the effects of particular oncogenes. It is possible to generate transgenic buds by using RCAS vectors to infect cells that contribute to the germline. However, the most common use of the RCAS system in chickens is to explore the effects of the expression of individual genes on embryonic development. In the simplest form of these experiments, the virus is introduced into a particular tissue or organ and allowed to spread. If the size of the inoculum and the duration of the experiment are appropriately chosen, the effects of the RCAS vector — and, more particularly, the gene it carries — are confined to the desired tissue or organ. If this is not sufficient, the spread of the virus can be confined using surgically constructed chimeric embryos. Not all strains of chickens express functional receptors for all the ASLVs. If a chimeric embryo is constructed from tissues from a permissive and a nonpermissive embryo, the virus will be confined to those tissues derived from the permissive embryo (Fekete and Cepko, 1993). It is also possible to infect chicken embryos with replication-defective versions of RCAS. If the stocks of the replication-defective vector are carefully prepared (i.e., free of replication-competent helper virus), the vector will not spread in the embryo. This system can also be used to follow cell fate/lineage in a developing embryo.
In thinking about experiments infecting embryos with RCAS vectors, it is worth considering the use of infected cells instead of virus. Infected cells are an excellent source of virus and can, at least in some cases, be delivered with more precision and in a smaller volume than a viral stock.

Ultimately, we would like to be able to provide a comprehensive set of vectors. In effect, if someone were to write down the properties of a vector — which cells or tissues it would infect, which cells or tissues it would replicate in, which cells or tissues it would be expressed in, etc. — then we could supply the relevant vector. Progress has been made, but there are some problems that have not been solved. In some cases, we know how we should proceed but haven’t had sufficient time to build the right vectors. In other cases, additional research will be required, and we are not certain when, if ever, the problems will be fixed. Here is a short list of some of the things that we would like to do:
- Although we initially planned to produce helper cells for BBAN, we have found that complementation with VSV-G in transient transfections works quite well, and there is no replication-competent virus produced.
- Make an RCAS derivative that will replicate efficiently in mammalian cells. As we said, the fact that RCAS vectors cannot replicate in mammalian cells has some advantages, but also has some disadvantages. What we would really like is to have a choice: RCAS vectors that can replicate on mammalian cells as well as those that cannot. This has been a difficult problem. Although we’ve made considerable progress, we do not yet have a vector with all of the desired properties.
- Although MLV cannot infect nondividing cells, it was recently shown that ASLVs (including RCAS vectors) can infect nondividing cells in culture, albeit at a lower efficiency relative to infection of dividing cells (Hatziioannou and Goff, 2001 Katz et al., 2002). We are exploring the possibility that RCAS vectors may be useful for infecting nondividing cells in animal models.
- Create modified envelope genes that will direct the RCAS vectors to the cell surface protein of our choice. This is the holy grail of retroviral vector design. We’re working on this particularly difficult problem, as are a number of other laboratories. We hope that some of the recent advances in understanding the structure and function of retroviral envelope genes will help solve this problem.
- Having developed both a shuttle vector (RSVP) and a gene-trap vector (pGT-GFP), we are trying to create a shuttle-trap vector that has the advantages of both.

The parental ASLV vectors are doubly defective in mammalian cells: the viruses cannot efficiently enter mammalian cells; if this barrier is overcome, mammalian cells do not release infectious ASLV particles. We have resolved the first of these two problems and are now working on the second. Basically, the problem with entry is that mammalian cells lack functional receptors for any of the standard ASLV envelope glycoproteins. This limitation can be overcome in two ways. First, it is possible to introduce a cloned ASLV receptor into mammalian cells, which can then be infected by RCAS vectors having the cognate envelope glycoprotein (Bates et al., 1993). Second, it is also possible to derive RCAS vectors that use the envelope glycoproteins normally found in murine retroviruses. These modified RCAS vectors will replicate in appropriate chicken cell lines, and can efficiently infect (but will not replicate in) mammalian cells that express the appropriate murine leukemia virus (MLV) receptors.
Although the fact that ASLVs cannot replicate in mammalian cells is not always useful, in some experiments the replication-defective nature of the vector is quite helpful. The defect is absolute; there are no problems with recombination with a helper virus when the vector stock is derived, nor are there problems with the vector recombining with or being mobilized by endogenous murine viruses, either in cultured mouse cells or in the animals themselves.
There are two different systems that can be used to introduce the RCAS vectors into mice. First, RCAS vectors with MLV envelopes can be used. Vectors of this type can be used with any of the available mouse strains or mouse mutants. The second system involves transgenic mice that express the tva receptor. The advantage of this system is that the distribution of the receptor can be controlled by the experimentalist. We have prepared two sets of transgenic mice. The first has the cDNA linked to a beta-actin promoter and expresses the receptor in all the tissues we tested. The second has the tva receptor linked to the alpha-actin promoter and expresses primarily in striated muscle. Other laboratories have produced mouse lines that express under a variety of tissue-specific promoters. This system makes it possible to direct an RCAS vector to a specific cell type or tissue type. Because it is both slow and expensive to prepare a new transgenic mouse line, we often suggest that people try the beta-actin tva mouse strain first. This strain can be used for preliminary experiments; if the preliminary experiments look promising, it is easier to justify the work of preparing a new transgenic line. The beta-actin tva line is also a good source of primary cells/cell lines that can be efficiently infected in culture. The original strains of tva transgenic mice were Black 6 backcross animals; we are now characterizing new transgenic lines prepared with pure Black 6 animals and are doing additional backcrosses with the original lines.
When we infect mice (or mouse blastocysts) with RCAS vectors, we usually use infected avian cells rather than a viral stock. The infected cells can be delivered in a small volume, and the subsequent infection of the mouse is more efficient if virally infected cells are used rather than a virus stock.
The following list provides an introduction to the literature pertaining to RCAS vectors:
- The RCAS vector system. Hughes, S.H. (2004) Folia Biologica 50: 107-119. [Full-text PDF]
- Retroviral gene delivery. Federspiel, M.J., and Hughes, S.H. (1997) Methods Cell Biol. 52: 179-214. [Chapter 9: Retroviral Gene Delivery]
- PubMed listing of articles related to the above review by Federspiel and Hughes
- Selected publications on retroviral vectors by the Hughes laboratory
- Retroviruses. Coffin, J.M., Hughes, S.H., and Varmus, H.E., editors. (1997) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Full-text online book on National Library of Medicine website]
- References section of Chapter 9: Development and Applications of Retroviral Vectors in Retroviruses
- How can I obtain DNA for the RCAS vectors and adaptors?
If you are at a nonprofit research laboratory and have no commercial interest in the RCAS system, we can help you execute an NIH simple letter agreement (SLA). Once the SLA forms are completed, we will ship the plasmid DNA to you. If you are at a commercial organizations or have any commercial interests, you must negotiate directly with the Technology Transfer Center, National Cancer Institute. - How can I obtain DF-1 cells?
The permanent EV-0-derived chicken cell line DF-1 was prepared by Doug Foster (University of Minnesota) and can be obtained from the American Type Culture Collection (ATCC; catalog #CRL-12203). Note that this cell line is not listed in the ATCC website, but is available through telephone orders (800-638-6597 or 703-365-2700). - How can I obtain transgenic mice that express tva?
This requires both an SLA (see FAQ #1) and official assurances for animal welfare. Shipping the animals can be expensive (particularly to Europe). We can help with the paperwork; anyone with a commercial interest will have to negotiate with the TDCB, NCI. - Where can I get the sequence of the vector?
We have provisional sequences (downloadable as text files) for several of the commonly used RCAS vectors: RCASBP(A), RCASBP(B), and RCASBP(M). Numbering of these sequences begins at the U3 R boundary in the lefthand LTR. Note that the sequences have not been finalized. We are in the process of preparing the final versions of the sequences. As soon as this has been done, the corrected sequences will be posted here. - If the ClaI cloning site is unique, why does the sequence have two ClaI sites listed?
As strange as it must seem now, the original RCAS cleavage site map was determined by digestion with restriction enzymes, not from the sequence. The upstream ClaI site is subject to Dam methylation in E. coli, and is not cleaved if the plasmid DNA is grown in a Dam(+) E. coli strain. - What RCAS vectors are available?
This website should give you a reasonable idea of the vectors we can supply. If you want something exotic, we probably can help you find what you need, or give you the materials you need to construct the vector you want. - I’m having trouble putting inserts into the ClaI site — What’s the problem? Either bad DNA or bad ClaI. There doesn’t seem to be any restriction on putting inserts into the ClaI site in the RCAS plasmids and cloning the plasmid in E. coli.; ClaI is not the most reliable restriction enzyme; we’ve been sold bad batches. If the digested DNA looks fine on a gel but won’t ligate, the ClaI probably contains some 3' DNA exonuclease, a problem that was much worse 5–10 years ago. If you are having such problems, check your ligation reaction before you do the transformation.
- I don't get any virus back after the cells are transfected — What's wrong? Basically, if you use a simple transfection protocol to put 5-10 micrograms of RCAS DNA onto a plate of compatible chicken cells, you will get virus back. If you are using CEFs, make sure the cells have the receptor for the envelope you have chosen. Also, make sure your cells are not contaminated with an ASLV that will block the replication of the RCAS vector.
- I get back replicating virus, but the insert is gone — What's wrong? Unfortunately, there are a number of possibilities; refer to Overview — What to Avoid in Vector Design.
- My institutional biosafety committee (IBC) wants to know how I plan to handle the vectors — What do you do? If you use the RCAS vectors with avian envelopes, the viruses will not infect mammalian cells. Even the versions that have murine envelopes, which can infect mammalian cells, do not replicate in mammalian cells. That having been said, we recommend that the vectors be handled carefully because they are infections agents. We handle the viruses with avian envelopes under BL-1 condition and those with mammalian envelopes under BL-2
The following list provides links to information related to RCAS vectors*:
- Plasmid: RCAS BP (A). Vector Database (a digital collection of vector backbones compiled by the nonprofit plasmid repository Addgene as a free informational resource for the scientific community).
- Somatic genome editing with the RCAS-TVA-CRISPR-Cas9 system for precision tumor modeling. Oldrini, B., Curiel-García, Á., Marques, C., Matia, V., Uluçkan, Ö., Graña-Castro, O., Torres-Ruiz, R., Rodriguez-Perales, S., Huse, J.T., and Squatrito, M. (2018) Nat. Commun. 9: 1466, doi: 10.1038/s41467-018-03731-w. [Full-text PDF]
- Model of the TVA receptor determinants required for efficient infection by subgroup A avian sarcoma and leukosis viruses. Melder, D.C., Pike, G.M., VanBrocklin, M.W., and Federspiel, M.J. (2015) J. Virol. 89: 2136-2148, doi: 10.1128/JVI.02339-14. [Full-text PDF]
- Using the RCAS-TVA system to model human cancer in mice. Ahronian, L.G., and Lewis, B.C. (2014) Cold Spring Harb. Protoc. 2014(11): 1128-1135, doi:10.1101/pdb.top069831. [Full-text PDF]
- Generation of high-titer RCAS virus from DF1 chicken fibroblasts. Ahronian, L.G., and Lewis, B.C. (2014) Cold Spring Harb. Protoc. 2014(11): 1161-1166, doi:10.1101/pdb.prot077974. [Full-text PDF]
- In vivo delivery of RCAS virus to mice. Ahronian, L.G., and Lewis, B.C. (2014) Cold Spring Harb. Protoc. 2014(11): 1167-1169, doi:10.1101/pdb.top069831. [Full-text PDF]
- Production of avian retroviruses and tissue-specific somatic retroviral gene transfer in vivo using the RCAS/TVA system. von Werder, A., Seidler, B., Schmid, R.M., Schneider, G., and Saur, D. (2012) Nat. Protoc. 7: 1167-1183, doi: 10.1038/nprot.2012.060. [Full-text PDF]
- RCAS-TVA in the mammary gland: An in vivo oncogene screen and a high fidelity model for breast transformation? Du, Z., and Li, Y. (2007) Cell Cycle 6: 823-826, doi: 10.4161/cc.6.7.4074. [Full-text PDF]
- An RCAS-TVA-based approach to designer mouse models. Orsulic, S. (2002) Mamm. Genome 13: 543-547, doi: 10.1007/s00335-002-4003-4. [Full-text PDF]
Requests
This site is intended to help you understand what reagents would be useful in your experiments. Because there are many choices, please be specific when requesting reagents from us. Also, refer to the Frequently Asked Questions (FAQs) section for additional information about what needs to be done under certain circumstances to fulfill your requests.
Mary Kearney, Ph.D.
Deputy Director
HIV Dynamics and Replication Program
National Cancer Institute
NCI-Frederick, P.O. Box B, Bldg. 535 Rm 310
Frederick, MD 21702-1201
Phone: 301-846-6796
E-mail: kearneym@mail.nih.gov
Contact Us
For questions or comments about the RCAS system, please contact:
Stephen H. Hughes, Ph.D.
NIH Scientist Emeritus
HIV Dynamics and Replication Program
National Cancer Institute
NCI-Frederick, P.O. Box B, Building 535
Frederick, MD 21702-1201
Phone: 301-846-1619
E-mail: hughesst@mail.nih.gov