Kumaran S. Ramamurthi, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 37, Room 5132
- Bethesda, MD 20892
- 240-760-7881
- ramamurthiks@mail.nih.gov
RESEARCH SUMMARY
Dr. Ramamurthi’s laboratory studies fundamental mechanisms that cells use to differentiate and divide in an effort to understand how these processes may go awry during disease. His lab focuses on how proteins localize to particular subcellular locations and how they subsequently assemble to form large structures during development and cell division. Recently, he discovered that the shape of cellular membranes, either convex or concave, may recruit certain membrane shape-sensing proteins to their correct destination, a novel mechanism for subcellular protein localization.
Areas of Expertise

Kumaran S. Ramamurthi, Ph.D.
Research
Our laboratory employs classical genetics and biochemistry combined with fluorescence microscopy and biophysical and computational techniques to study how proteins localize in a cell and subsequently assemble into larger structures. We focus on protein localization events during cell division and morphogenesis.
Bacterial spore coat assembly as a model for understanding how large cellular structures are built. Spores are specialized bacterial cells that can remain dormant for decades and are highly resistant to environmental insults. Spores are encased in a thick protein shell termed the “coat”. The basement layer of the coat contains two proteins that initiate coat assembly. SpoVM is a tiny protein that anchors the coat to the spore’s surface. We discovered that SpoVM localizes during spore formation by preferentially embedding in micron-scale convex (positively curved) membranes such as that found on the spore’s surface. SpoVM then recruits SpoIVA, a novel cytoskeletal protein, that we discovered hydrolyzes ATP to irreversibly polymerize to form a shell that encases the developing spore. We are currently studying how later steps in coat assembly proceed and how coat assembly is monitored.
Assembly of synthetic bacteria to deliver drugs. Recently, we have reconstituted the assembly of the basement layer of the spore coat, using purified SpoVM and SpoIVA, around bacteria-sized silica beads surrounded by a membrane to construct synthetic spore-like particles that we named “SSHELs”. We are currently investigating the utility of SSHEL particles in delivering chemotherapeutics specifically to tumors.
Regulation of cellular differentiation. Bacterial spore formation (called “sporulation”) is a relatively simple developmental pathway in which a progenitor cell asymmetrically divides, then irreversibly commits to creating two genetically identical daughter cells that exhibit different cell fates (one daughter becomes a spore, and the other daughter nurtures the developing spore and eventually lyses). Entry into sporulation is governed by several well-studied checkpoints, but later steps that regulate progression through sporulation are less well understood. We are currently identifying quality control pathways that govern progression through sporulation after the cell commits to completing this developmental program.
Bacterial cell division. Correct placement of the cell division septum is a fundamental feature of life. We are currently identifying and characterizing components that determine the asymmetric placement of the division septum in the rod-shaped model bacterium Bacillus subtilis when it initiates the differentiation pathway that culminates in the formation of a spore. We are also investigating how a spherical bacterium, the human pathogen Staphylococcus aureus, correctly identifies mid-cell and regulates cell division.
Publications
Altruistic feeding and cell-cell signaling during bacterial differentiation actively enhance phenotypic heterogeneity
Bacterial spore surface nanoenvironment requires a AAA+ ATPase to promote MurG function
Biography

Kumaran S. Ramamurthi, Ph.D.
Kumaran Ramamurthi received his Ph.D. in Molecular Biology from the University of California, Los Angeles (UCLA), where he studied the secretion of bacterial virulence proteins. After a brief research fellowship at the University of Chicago, he studied subcellular protein localization as a Ruth L. Kirchstein National Research Service Award Postdoctoral Fellow at Harvard University.
He arrived at the NIH in 2009 as a Tenure Track Investigator and was promoted to Senior Investigator in 2016. He currently serves on the Editorial Boards of The Journal of Biological Chemistry and Microbiology and Molecular Biology Reviews. He received the National Cancer Institute Director’s Award in 2016 and was elected to the American Academy of Microbiology in 2025.
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Team
News
Covers

Cell-specific cargo delivery using synthetic bacterial spores
Kong et al. use synthetic bacterial spores, called “SSHELs,” that deliver doxorubicin to HER2-positive ovarian tumors. Target cells actively internalize bound SSHELs and traffic them to lysosomes, where the acidic pH liberates the drug. Depicted is a nautical interpretation of this interplay between ovarian cancer cells and SSHEL particles, wherein SSHELs (yellow) plunge into a wave, which represents the surface of a target cell undergoing macropinocytosis, to uptake the particles. Upon entry, SSHELs unleash the enclosed cargo (red) into the oceanic cytosol. Artwork by Erina He, NIH Medical Arts.
Kong M, D'Atri D, Bilotta MT, Johnson B, Updegrove TB, Gallardo DL, Machinandiarena F, Wu IL, Constantino MA, Hewitt SM, Tanner K, Fitzgerald DJ, Ramamurthi KS. Cell Rep. 2023 Jan 31;42(1):111955. doi: 10.1016/j.celrep.2022.111955. Epub 2023 Jan 4.PMID: 36640333
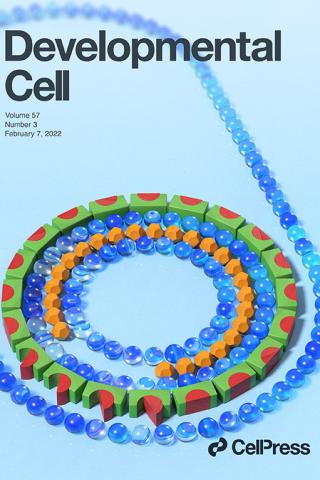
Bacterial developmental checkpoint that directly monitors cell surface morphogenesis
The bacterium Bacillus subtilis undergoing spore formation is depicted using toys. Membranes are depicted using marbles. The spore is encased in two concentric shells: an outer proteinaceous “coat” (green and red blocks) and a peptidoglycan “cortex” (orange dodecahedron). To learn more about how coat assembly is linked to cortex formation, see Delerue et al. (pp. 344–360).
Delerue, T., Anantharaman, A., Gilmore, M.C., Popham, D.L., Cava, F., Aravind, L., and Ramamurthi, K.S. Bacterial developmental checkpoint that directly monitors cell surface morphogenesis Developmental Cell 2022; 57:344-360 <https://www.sciencedirect.com/science/article/pii/S1534580721010418?via%3Dihub>

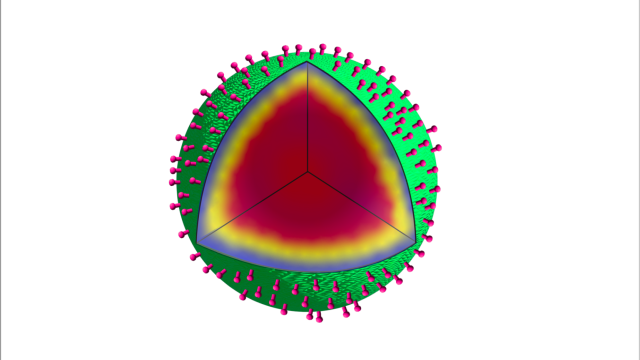
Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis
The Min system in rod-shaped bacteria restricts improper assembly of the division septum. In Escherichia coli, the Min system localizes to the cell poles, but in Bacillus subtilis, it is recruited to nascent cell division sites at mid-cell to prevent aberrant septation events immediately adjacent to a constricting septum. How does the cell spatially and temporally restrict the inhibitory activity of the Min system so that it does not interfere with normal cell division? This image reveals (using a super-resolution fluorescence microscopy technique called SIM) that the cell division protein DivIVA (green), which preferentially localizes to negatively curved membranes (red) and is responsible for recruitment of the Min system, localizes as double rings on either side of actively constricting septa and remains associated with mature septa after completion of cell division.
In the related article, the authors propose that DivIVA interprets membrane invagination as evidence of cell division and localizes to mid-cell only after the onset of membrane constriction. Additionally, the formation of two ring-shaped platforms on either side of the septum reveals a mechanism by which the inhibitory activity of the Min system is held away from a newly forming septum, while simultaneously inhibiting aberrant septation at sites immediately adjacent to mid-cell.
Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis
Prahathees Eswaramoorthya, Marcella L. Erbb, James A. Gregoryb, Jared Silvermanc, Kit Poglianob, Joe Poglianob, and Kumaran S. Ramamurthia. mBio 2(6):e00257–11, 2011.
aLaboratory of Molecular Biology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA
bDivision of Biological Sciences, University of California at San Diego, La Jolla, California, USA
cCubist Pharmaceuticals, Lexington, Massachusetts, USA