Xue Zhi Zhao, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Bldg. 376, Rm. 217
- Frederick, MD 21702-1201
- 301-846-5907
- xuezhi.zhao@nih.gov
RESEARCH SUMMARY
Dr. Zhao’s research focuses on the design and development of target-directed small molecules and peptides for therapeutic applications. Using state-of-the-art medicinal chemistry, biochemical, pharmacological, and molecular biology techniques, he leads innovative drug discovery efforts targeting key disease-related proteins, including integrase (IN), polo-like kinase 1 (Plk1), and tyrosyl-DNA phosphodiesterase 1 (TDP1). His work includes the development of integrase strand transfer inhibitors (INSTIs) with potent activity against both wild-type and drug-resistant integrase variants, as well as highly selective peptide ligands for the Plk1 polo-box domain. He has also pioneered a novel class of TDP1 inhibitors designed to enhance the efficacy of topoisomerase I-based cancer therapies. By integrating structural biology, fragment-based screening, and advanced synthetic strategies such as oxime diversification and multicomponent reactions, Dr. Zhao continues to advance promising drug candidates with strong translational potential.
Areas of Expertise
Research
Dr. Zhao's research focuses on the design and development of small-molecule or peptide therapeutics targeting key enzymes involved in viral replication and cancer progression.
Integrase Strand Transfer Inhibitors (INSTIs)
Multiple structurally distinct classes of INSTIs have been developed, including 2,3-dihydroxybenzoylhydrazides, isoindolones, sulfonamides, and pyrrolopyridine-trione derivatives. These compounds exhibit potent low-nanomolar inhibition of integrase, strong antiviral activity, and reduced cytotoxicity. A recent series of 4-amino-1-hydroxy-1,8-naphthyridine-3-carboxamides demonstrated exceptional efficacy against both wild-type and drug-resistant variants, surpassing the currently approved INSTIs. Cryo-EM structures of lead compounds bound to the intasome are guiding further structure-based optimization. Preclinical pharmacokinetic and formulation studies, including non-human primate evaluations, are ongoing in collaboration with the NCI Invention Development Program.
Tyrosyl-DNA Phosphodiesterase 1 (TDP1) Inhibitors
The development of TDP1 inhibitors has resulted in a novel class of compounds intended to synergize with topoisomerase I (TOP1) inhibitors in cancer therapy. Fragment-based X-ray crystallography and high-throughput small molecule microarray screening identified key scaffolds, including phthalic acid derivatives and diphenylimidazo[1,2-a]pyrazines. Further optimization via multicomponent synthesis and oxime diversification produced multivalent inhibitors phenylimidazo[1,2-a]pyridines capable of engaging DNA, protein, and catalytic sites within TDP1. These compounds represent promising leads for future therapeutic development.
Polo-like Kinase 1 (Plk1) Polo-Box Domain Peptide Inhibitors
Oxime diversification and click chemistry have been applied to design high-affinity peptide ligands targeting a cryptic pocket in the Plk1 polo-box domain. These ligands exhibit significantly enhanced binding affinity and selectivity, supported by the identification of an auxiliary binding region adjacent to a key hydrophobic site. This has enabled improved discrimination of Plk1 over closely related Plk2 and Plk3 polo-box domains.
Publications
- Bibliography Link
- View Dr. Zhao's Complete Bibliography at NCBI.
- View Dr. Zhao's ORCID page.
Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Biography

Xue Zhi Zhao, Ph.D.
Dr. Zhao is a Senior Associate Scientist in the Chemical Biology Laboratory at the National Cancer Institute. He earned his Ph.D. in organic chemistry from Lanzhou University and joined the NCI as a NIH Visiting Fellow, progressing through roles as Research Fellow, Staff Scientist, and Associate Scientist. Dr. Zhao specializes in the design of target-directed small molecules and peptides for therapeutic applications. His research focuses on the development of integrase strand transfer inhibitors (INSTIs) to combat drug-resistant viral strains, selective peptide inhibitors of the polo-like kinase 1 (Plk1) polo-box domain, and tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors to enhance topoisomerase I-based cancer therapies. By integrating medicinal chemistry, structural biology, and advanced synthetic strategies, Dr. Zhao’s work continues to generate promising candidates with strong potential for clinical translation.
Covers

Identification of multidentate tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors that simultaneously access the DNA, protein and catalytic-binding sites by oxime diversification
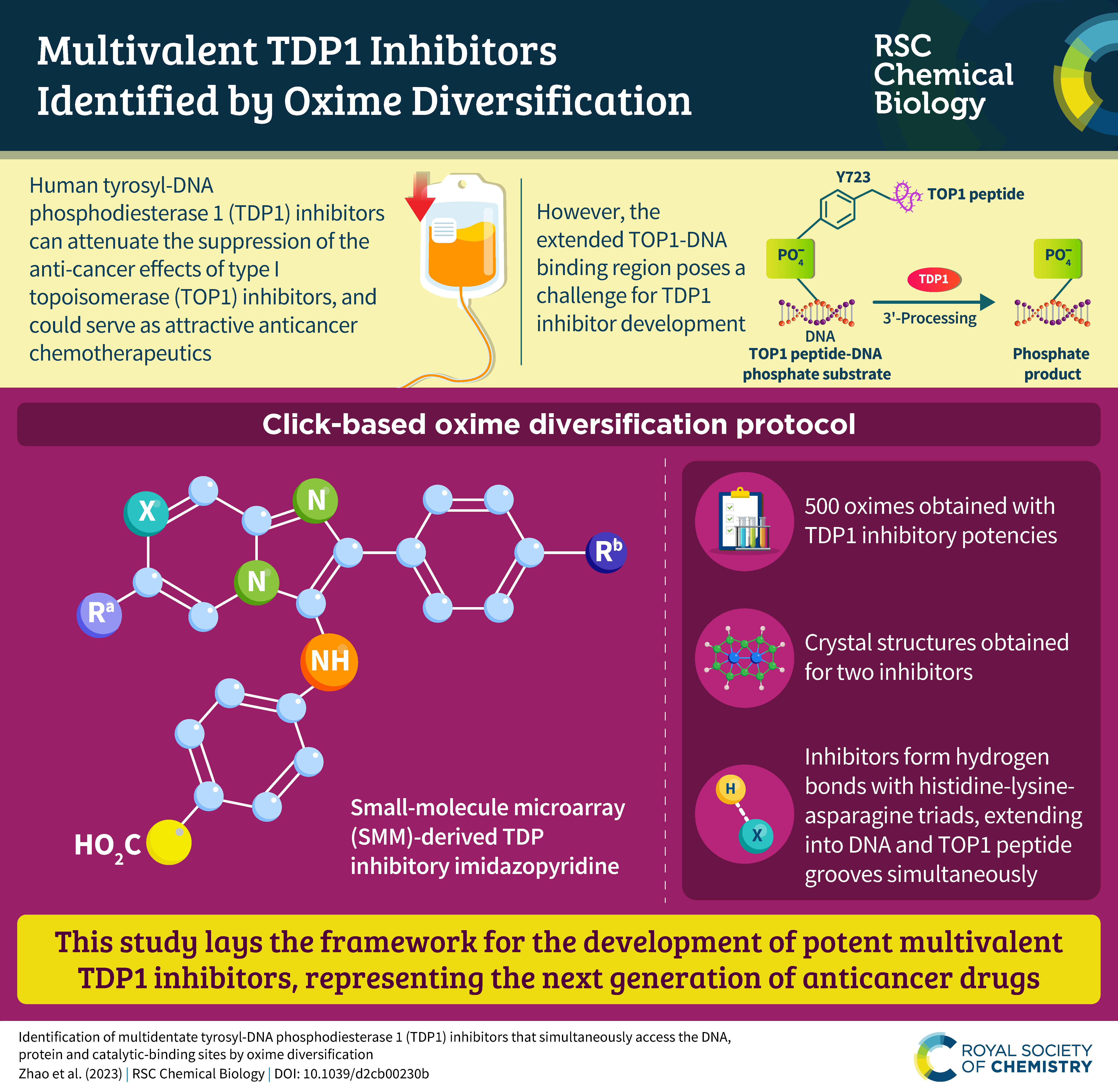
Tyrosyl-DNA phosphodiesterase 1 (TDP1) is a member of the phospholipase D family that can downregulate the anticancer effects of the type I topoisomerase (TOP1) inhibitors by hydrolyzing the 3'-phosphodiester bond between DNA and the TOP1 residue Y723 in the critical stalled intermediate that is the foundation of TOP1 inhibitor mechanism of action. Thus, TDP1 antagonists are attractive as potential enhancers of TOP1 inhibitors. However, the open and extended nature of the TOP1–DNA substrate binding region has made the development of TDP1 inhibitors extremely challenging. In this study, starting from our recently identified small molecule microarray (SMM)-derived TDP1-inhibitory imidazopyridine motif, we employed a click-based oxime protocol to extend the parent platform into the DNA and TOP1 peptide substrate-binding channels. We applied one-pot Groebke–Blackburn–Bienayme multicomponent reactions (GBBRs) to prepare the needed aminooxy-containing substrates. By reacting these precursors with approximately 250 aldehydes in microtiter format, we screened a library of nearly 500 oximes for their TDP1 inhibitory potencies using an in vitro florescence-based catalytic assay. Select hits were structurally explored as their triazole- and ether-based isosteres. We obtained crystal structures of two of the resulting inhibitors bound to the TDP1 catalytic domain. The structures reveal that the inhibitors form hydrogen bonds with the catalytic His-Lys-Asn triads (‘‘HKN’’ motifs: H263, K265, N283 and H493, K495, N516), while simultaneously extending into both the substrate DNA and TOP1 peptide-binding grooves. This work provides a structural model for developing multivalent TDP1 inhibitors capable of binding in a tridentate fashion with a central component situated within the catalytic pocket and extensions that project into both the DNA and TOP1 peptide substrate-binding regions.
Xue Zhi Zhao, Wenjie Wang, George T. Lountos, Evgeny Kiselev, Joseph E. Tropea, Danielle Needle, Yves Pommier and Terrence R. Burke, Jr.
HIV‑1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants
Integrase strand transfer inhibitors (INSTIs) block the integration step of the retroviral lifecycle and are first-line drugs used for the treatment of HIV-1/AIDS. INSTIs have a polycyclic core with heteroatom triads, chelate the metal ions at the active site, and have a halobenzyl group that interacts with viral DNA attached to the core by a flexible linker. The most broadly effective INSTIs inhibit both wild-type (WT) integrase (IN) and a variety of well-known mutants. However, because there are mutations that reduce the potency of all the available INSTIs, new and better compounds are needed. Models based on recent structures of HIV-1 and red-capped mangabey SIV INs suggest modifications in the INSTI structures that could enhance interactions with the 3′-terminal adenosine of the viral DNA, which could improve performance against INSTI resistant mutants. We designed and tested a series of INSTIs having modifications to their naphthyridine scaffold. One of the new compounds retained good potency against an expanded panel of HIV-1 IN mutants that we tested. Our results suggest the possibility of designing inhibitors that combine the best features of the existing compounds, which could provide additional efficacy against known HIV-1 IN mutants.
Steven J. Smith, Xue Zhi Zhao, Dario Oliveira Passos, Valerie E. Pye, Peter Cherepanov, Dmitry Lyumkis, Terrence R. Burke, Jr., and Stephen H. Hughes

Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
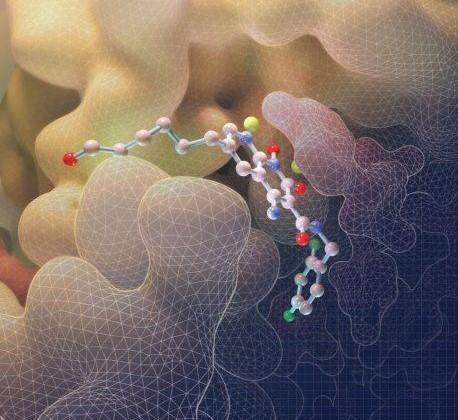
We now report examining a 21 000-member library of drug-like Small Molecules in Microarray (SMM) format for their ability to bind Alexa Fluor 647 (AF647)-labeled Tyrosyl-DNA phosphodiesterase 1 (TDP1). The screen identified structurally similar N,2-diphenylimidazo[1,2-a]pyrazin-3-amines as TDP1 binders and catalytic inhibitors. We then explored the core heterocycle skeleton using one-pot Groebke–Blackburn–Bienayme multicomponent reactions and arrived at analogs having higher inhibitory potencies. Solving TDP1 co-crystal structures of a subset of compounds showed their binding at the TDP1 catalytic site, while mimicking substrate interactions. Although our original fragment screen differed significantly from the current microarray protocol, both methods identified ligand–protein interactions containing highly similar elements. Importantly inhibitors identified through the SMM approach show competitive inhibition against TDP1 and access the catalytic phosphate-binding pocket, while simultaneously providing extensions into both the substrate DNA and peptide-binding channels. As such, they represent a platform for further elaboration of trivalent ligands, that could serve as a new genre of potent TDP1 inhibitors.
Xue Zhi Zhao, Evgeny Kiselev, George T. Lountos, Wenjie Wang, Joseph E. Tropea, Danielle Needle, Thomas A. Hilimire, John S. Schneekloth, Jr, David S. Waugh, Yves Pommier and Terrence R. Burke, Jr.

Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening
Tyrosyl DNA-phosphodiesterase I (TDP1) repairs type IB topoisomerase (TOP1) cleavage complexes generated by TOP1 inhibitors commonly used as anticancer agents. TDP1 also removes DNA 3′ end blocking lesions generated by chain-terminating nucleosides and alkylating agents, and base oxidation both in the nuclear and mitochondrial genomes. Combination therapy with TDP1 inhibitors is proposed to synergize with topoisomerase targeting drugs to enhance selectivity against cancer cells exhibiting deficiencies in parallel DNA repair pathways. A crystallographic fragment screening campaign against the catalytic domain of TDP1 was conducted to identify new lead compounds. Crystal structures revealed two fragments that bind to the TDP1 active site and exhibit inhibitory activity against TDP1. These fragments occupy a similar position in the TDP1 active site as seen in prior crystal structures of TDP1 with bound vanadate, a transition state mimic. Using structural insights into fragment binding, several fragment derivatives have been prepared and evaluated in biochemical assays. These results demonstrate that fragment-based methods can be a highly feasible approach toward the discovery of small-molecule chemical scaffolds to target TDP1, and for the first time, we provide co-crystal structures of small molecule inhibitors bound to TDP1, which could serve for the rational development of medicinal TDP1 inhibitors.
George T. Lountos, Xue Zhi Zhao, Evgeny Kiselev, Joseph E. Tropea, Danielle Needle, Yves Pommier, Terrence R. Burke, Jr. and David S. Waugh

Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors
Integrase mutations can reduce the effectiveness of the first-generation FDA-approved integrase strand transfer inhibitors (INSTIs), raltegravir (RAL) and elvitegravir (EVG). The second-generation agent, dolutegravir (DTG), has enjoyed considerable clinical success; however, resistancecausing mutations that diminish the efficacy of DTG have appeared. Our current findings support and extend the substrate envelope concept that broadly effective INSTIs can be designed by filling the envelope defined by the DNA substrates. Previously, we explored 1-hydroxy-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamides as an INSTI scaffold, making a limited set of derivatives, and concluded that broadly effective INSTIs can be developed using this scaffold. Herein, we report an extended investigation of 6-substituents as well the first examples of 7-substituted analogues of this scaffold. While 7-substituents are not well-tolerated, we have identified novel substituents at the 6-position that are highly effective, with the best compound (6p) retaining better efficacy against a broad panel of known INSTI resistant mutants than any analogues we have previously described. View the article.
Xue Zhi Zhao, Steven J. Smith, Daniel P. Maskell, Mathieu Metífiot, Valerie E. Pye, Katherine Fesen, Christophe Marchand, Yves Pommier, Peter Cherepanov, Stephen H. Hughes, and Terrence R. Burke, Jr.

HIV‑1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases
On the cover: Mutant forms of HIV-1 IN reduce the therapeutic effectiveness of integrase strand transfer inhibitors (INSTIs). The cover figure shows the IN of prototype foamy virus complexed to a novel INSTI (gold) that retains potency against resistant mutants of HIV-1 IN. Overlain are the host and viral DNA substrates (blue and green, respectively), showing substrate mimicry by the inhibitor.
Xue Zhi Zhao, Steven J. Smith, Daniel P. Maskell, Mathieu Metifiot, Valerie E. Pye, Katherine Fesen, Christophe Marchand, Yves Pommier, Peter Cherepanov, Stephen H. Hughes and Terrence R. Burke, Jr.

Application of oxime-diversification to optimize ligand interactions within a cryptic pocket of the polo-like kinase 1 polo-box domain
By a process involving initial screening of a set of 87 aldehydes using an oxime ligation-based strategy, we were able to achieve a several-fold affinity enhancement over one of the most potent previously known polo-like kinase 1 (Plk1) polo-box domain (PBD) binding inhibitors. This improved binding may result by accessing a newly identified auxiliary region proximal to a key hydrophobic cryptic pocket on the surface of the protein. Our findings could have general applicability to the design of PBD-binding antagonists.
Cover design by Joseph Myer
Xue Zhi Zhao, David Hymel and Terrence R. Burke, Jr.
Bioorganic & Medicinal Chemistry Letters, 2016, 26, 5009-5012.
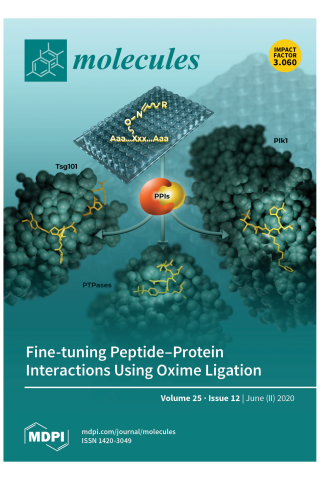
Application of Post Solid-Phase Oxime Ligation to Fine-Tune Peptide-Protein Interactions
Protein-protein interactions (PPIs) represent an extremely attractive class of potential new targets for therapeutic intervention; however, the shallow extended character of many PPIs can render developing inhibitors against them as exceptionally difficult. Yet this problem can be made tractable by taking advantage of the fact that large interacting surfaces are often characterized by confined "hot spot" regions, where interactions contribute disproportionately to overall binding energies. Peptides afford valuable starting points for developing PPI inhibitors because of their high degrees of functional diversity and conformational adaptability. Unfortunately, contacts afforded by the 20 natural amino acids may be suboptimal and inefficient for accessing both canonical binding interactions and transient "cryptic" binding pockets. Oxime ligation represents a class of biocompatible "click" chemistry that allows the structural diversity of libraries of aldehydes to be rapidly evaluated within the context of a parent oxime-containing peptide platform. Importantly, oxime ligation represents a form of post solid-phase diversification, which provides a facile and empirical means of identifying unanticipated protein-peptide interactions that may substantially increase binding affinities and selectivity. The current review will focus on the authors' use of peptide ligation to optimize PPI antagonists directed against several targets, including tumor susceptibility gene 101 (Tsg101), protein tyrosine phosphatases (PTPases) and the polo-like kinase 1 (Plk1). This should provide insights that can be broadly directed against an almost unlimited range of physiologically important PPIs.
Xue Zhi Zhao, Fa Liu and Terrence R. Burke Jr.
Molecules, 2020, 25 (12), 2807.
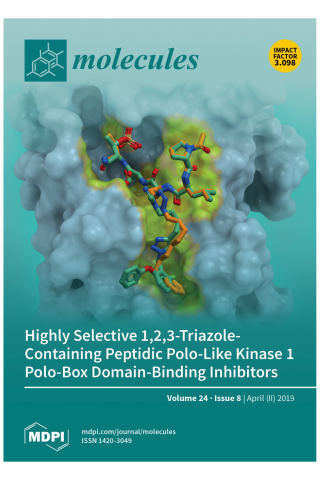
Development of Highly Selective 1,2,3-Triazole-containing Peptidic Polo-like Kinase 1 Polo-box Domain-binding Inhibitors
Cover Story: Two highly selective triazole-containing peptidic polo-like kinase 1 (Plk1) polo-box domain (PBD)-binding inhibitors (3d in green and 4b in orange) are depicted, binding within the hydrophobic “cryptic” binding pocket on the surface of the Plk1 PBD (light blue). The placement was guided by our previously reported X-ray crystal structure of PBD-bound parent peptide (2a) (PDB accession code: 3RQ7), whose binding pocket is highlighted in yellow. These ligands retain the high PBD-binding affinity of the parent peptide, while showing desirable enhanced selectivity for the PBD of Plk1 relative to the PBDs of Plk2 and Plk3.
Members of the polo-like kinase (Plk) family of serine/threonine protein kinases play crucial roles in cell cycle regulation and proliferation. Of five Plks (Plk1 – 5), Plk1 is recognized as an anticancer drug target. Plk1 contains multiple structural components that are important for its proper biological function. These include an N-terminal catalytic domain and a C-terminal non-catalytic polo-box domain (PBD). The PBD binds to phosphothreonine (pT) and phosphoserine-containing sequences. Blocking PBD-dependent interactions offers a potential means of down-regulating Plk1 function that is distinct from targeting its ATP-binding site. Previously, we demonstrated by tethering alkylphenyl chains from the N(π)-position of the His residue in the 5-mer PLHSpT, that we were able to access a hydrophobic "cryptic" binding pocket on the surface of the PBD, and in so doing enhance binding affinities by approximately 1000-fold. More recently, we optimized these PBD-ligand interactions using an oxime ligation-based strategy. Herein, using azide-alkyne cycloaddition reactions, we explore new triazole-containing PBD-binding antagonists. Some of these ligands retain the high PBD-binding affinity of the parent peptide, while showing desirable enhanced selectivity for the PBD of Plk1 relative to the PBDs of Plk2 and Plk3.
Xue Zhi Zhao, Kohei Tsuji, David Hymel and Terrence R. Burke Jr.
Molecules 2019, 24, 1488
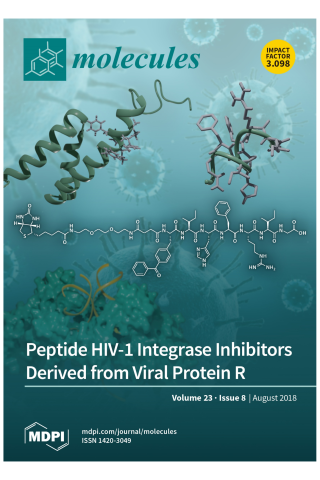
HIV-1 Integrase-Targeted Short Peptides Derived from a Viral Protein R Sequence
HIV-1 integrase (IN) inhibitors represent a new class of highly effective anti-AIDS therapeutics. Current FDA-approved IN strand transfer inhibitors (INSTIs) share a common mechanism of action that involves chelation of catalytic divalent metal ions. However, the emergence of IN mutants having reduced sensitivity to these inhibitors underlies efforts to derive agents that antagonize IN function by alternate mechanisms. Integrase along with the 96-residue multifunctional accessory protein, viral protein R (Vpr), are both components of the HIV-1 pre-integration complex (PIC). Coordinated interactions within the PIC are important for viral replication. Herein, we report a 7-mer peptide based on the shortened Vpr (69–75) sequence containing a biotin group and a photo-reactive benzoylphenylalanyl residue, and which exhibits low micromolar IN inhibitory potency. Photo-crosslinking experiments have indicated that the peptide directly binds IN. The peptide does not interfere with IN-DNA interactions or induce higher-order, aberrant IN multimerization, suggesting a mode of action for the peptide that is distinct from clinically used INSTIs and developmental allosteric IN inhibitors. This compact Vpr-derived peptide may serve as a valuable pharmacological tool to identify a potential new pharmacologic site.
Xue Zhi Zhao, Mathieu Métifiot, Evgeny Kiselev, Jacques J. Kessi, Kasthuraiah Maddali, Christophe Marchand, Mamuka Kvaratskhelia, Yves Pommier and Terrence R. Burke Jr.
Molecules, 2018, 23 (8), 1858.
.
Resources
‘Magic’ Frederick Collaboration Yields Encouraging Results
Through collaborative efforts, we examined a 21 000-member library of drug-like Small Molecules in Microarray (SMM) format for their ability to bind Alexa Fluor 647 (AF647)-labeled Tyrosyl-DNA phosphodiesterase 1 (TDP1). Inhibitors identified through the SMM approach show competitive inhibition against TDP1 and access the catalytic phosphate-binding pocket, while simultaneously providing extensions into both the substrate DNA and peptide-binding channels. This work, which appeared in Chemical Science, disclose a platform for further elaboration of trivalent ligands, that could serve as a new genre of potent TDP1 inhibitors.
Imaging study of key viral structure shows how HIV drugs work at atomic level
XZ426 is a latest generation of INSTI that highlights how small changes in the integrase active site can have notable implications for drug binding and design and provide mechanistic insights into why a leading INSTI retains efficacy against a broad spectrum of drug-resistant variants. This work, which appeared in Science, provides insights that could help design or improve new treatments for HIV.
HIV intasome bound INSTIs
6V3K (XZ419) 6PUY (XZ426) 6PUZ (XZ446) 8FNQ (HIV IN E138K/G140A/Q148K-XZ426)
PFV intasome bound INSTIs
4BDY (XZ89) 4BDZ (XZ90) 4BE0 (XZ115) 4BE1 (XZ116) 4BE2 (XZ259) 5MMA (XZ379) 5FRM (XZ384) 5NO1 (XZ407) 5FRN (XZ419) 5MMB (XZ434) 7ADU (XZ440) 5FRO (XZ446) 7ADV (XZ447)
STLV intasome bound INSTI
HIV reverse transcriptase (RT) bound RNase H inhibitor
TDP1 bound inhibitors
6DJF (XZ502) 6DJG (XZ503) 6DJH (XZ515) 6DJ5 (XZ519) 6MYZ (XZ520) 6DJI (XZ522) 6N0O (XZ523) 6MZ0 (XZ530) 6DJJ (XZ532) 6N0R (XZ572) 6N0N (XZ574) 6N0D (XZ575) 6N17 (XZ577) 6N19 (XZ578) 6W7L (XZ632p) 6W4R (XZ633p) 6W7K (XZ634p) 6W7J (XZ635p) 8CW2 (XZ760) 8CVQ (XZ761) 7UFY (XZ766) 7UFZ (XZ768)
Structures of IN bound 2nd generation INSTIs (Cherepanov, P. Maertens, G.N. Lyumkis, D.)
3S3M (PFV IN-DTG) 3S3N (PFV S217H-DTG) 3S3O (PFV N224H-DTG) 6RWN (SIVrcm IN-DTG)
6RWM (SIVrcm IN-BIC) 6RWO (SIVrcm G140S/Q148H-BIC) 7OUH (STLV IN:B56-BIC)
6PUW (HIV IN-BIC) 8FN7 (HIV IN-DTG) 8FNN (HIV IN E138K/G140A/Q148K-DTG)