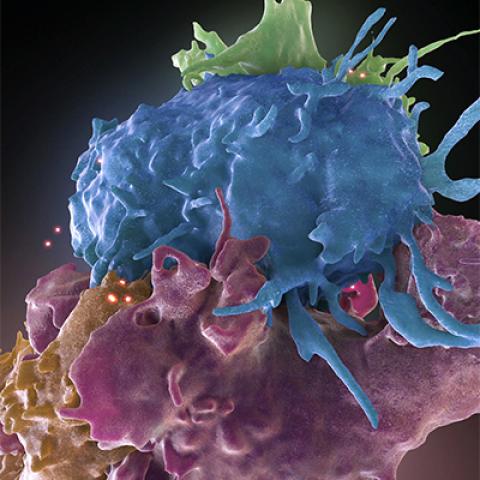
Three-dimensional structure of HIV infected (blue, green) and uninfected (brown, purple) T cells interacting. One cell (brown) has wrapped an extension around its uninfected neighbor (purple) to reach an infected cell (blue).
Image credit: Donald Bliss and Sriram Subramaniam
Even though antiretroviral therapies have allowed many people to live long lives, ridding the body of HIV completely has been an elusive goal ever since the discovery in the 1980s that HIV causes AIDS. CCR researchers, using advanced cellular interrogation techniques, have found that identical sequences contributing to HIV persistence are usually present due to one of three circumstances: the division and expansion of HIV-infected cells; a lack of proviral genetic expression (proviral latency); or the emergence of gene mutations due to drug resistance. The identical gene sequences can evolve prior to the start of antiretroviral therapy or later if the therapy is not successful at restraining viral replication.
Researchers have long known that CD4+ T cells are essential for an effective immune response to HIV. The mechanisms behind how clones of HIV-infected cells can survive for decades, however, despite the use of antiretroviral drugs that prevent viral spread, has remained elusive. Mary Kearney, Ph.D., Senior Scientist in the HIV Dynamics and Replication Program, and Sean Patro, Ph.D., a Postdoctoral Fellow, and colleagues, have advanced the understanding of proviral integration sites in a host cell’s DNA. Previously, scientists had not been able to link such sites with the populations of identical HIV proviruses that persist in patients on treatment.
In research reported in PNAS, Kearney and her colleagues used a newer technique, multiple-displacement amplification (MDA), on cells collected from blood and tissue of five patients who had been treated with antiretroviral therapy. Previously, researchers used two techniques developed in CCR, single-genome sequencing (SGS) and integration site analysis (ISA), to gain an understanding of the structure of persistent HIV DNA. But these tools did not provide evidence for how integration sites were linked to specific proviral sequences. MDA combines SGS and ISA to allow for sequencing of single HIV genomes and identification of their integration site in single cells, compounding the power of the two technologies to define the HIV reservoir. MDA also enabled the scientists to tease out ancestral HIV pieces from a repository of defective HIV-infected cells. Applying MDA techniques to analyzing HIV blood and tissue, Kearney says, could provide vital assistance to researchers trying to understand how HIV persists in the human body.
The researchers say that it will be very important to perform larger studies, especially those that identify and fully characterize the HIV genomes that not only persist on antiretroviral therapy but are the source of the recurrence of the infection if the therapy is interrupted. Kearney concludes that studies must be done on many more patients and must characterize many single HIV-infected cells within each patient, going beyond what was done in this initial pilot study. It is the researcher’s belief that larger studies will likely reveal targets that could be exploited to rid the body of HIV without antiretroviral therapy.