Michael Aregger, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 560, Room 12-92B
- Frederick, MD, 21702-1201
- 301-846-6093
- michael.aregger@nih.gov
RESEARCH SUMMARY
Michael Aregger’s research focuses on how cancer cells rewire gene expression and metabolism in order to adapt to changing environmental conditions. Towards this, he has developed CRISPR-Cas based screening technologies that afford the systematic perturbation of multiple genes and genetic segments and that can be coupled with various phenotypic readouts. Dr. Aregger’s laboratory applies those genome engineering tools and functional genomics approaches in order to reveal genetic interactions and cancer dependencies, and to identify regulators of metabolic plasticity in cancer cells.
Areas of Expertise

Michael Aregger, Ph.D.
Research
A major goal of biomedical research is to reveal genetic dependencies and cellular processes cancer cells rely on in order to exploit them therapeutically. The development of CRISPR-based genome editing technologies have opened the door for efficient perturbation of mammalian genomes and, as such, provide powerful tools for exploring genotype-to-phenotype relationships. Importantly, the combination of CRISPR technology with functional genomics approaches can be harnessed to systematically identify genetic dependencies and cancer vulnerabilities across diverse cell lines and growth conditions. Michael Aregger’s research aims to expand our understanding of how cancer cells dynamically regulate gene expression and cellular metabolism in response to changing environmental conditions, and to use this knowledge to identify genetic liabilities that may present novel targets for combinatorial cancer therapeutic approaches.
1. Studying adaptation to metabolic stress
A common feature of many cancers is the reprogramming of cellular metabolism in order to fuel tumorigenesis. Importantly, this rewiring is highly plastic allowing cancer cells to adapt to various tumor microenvironmental stress conditions. In our lab we apply cutting-edge functional genomics methodologies along with transcriptomics, proteomics and focused biochemical approaches to investigate how metabolic plasticity is regulated and what genetic vulnerabilities arise in cancer cells in response to various environmental stress conditions.
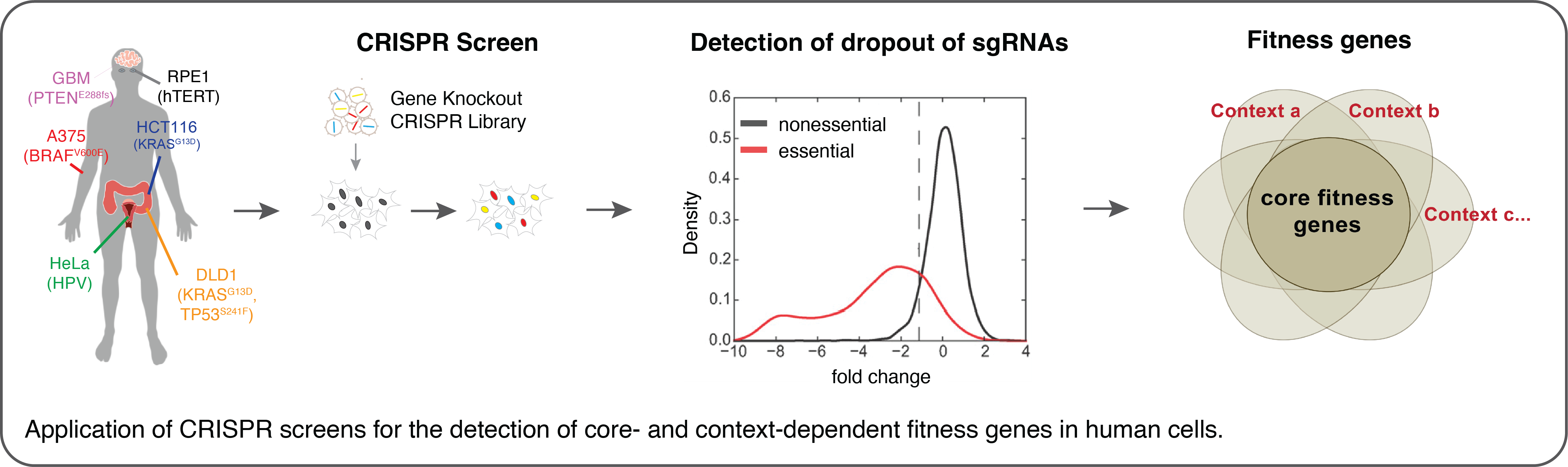
2. Identification of context-dependent fitness genes across genetic and environmental backgrounds
In order to identify genes whose function underlie cell proliferation and survival we have developed various CRISPR-based screening technologies for the systematic knockout of genes in human cells. Our genome-wide CRISPR-Cas9 screens expanded the set of human fitness genes and revealed distinct genetic signatures that can be used to predict differential drug response of cancer cells. In our lab we are using similar CRISPR screening approaches to uncover genetic vulnerabilities in cancer cells and study how different environmental conditions influence dependencies on certain cellular pathways.
3. Mapping of genetic interaction networks
Genetic interactions occur when mutations in two or more genes result in a phenotype that deviates from the predicted combinatorial effect of perturbing those genes. The mapping of such genetic interactions can provide profound insights into the functional wiring and plasticity of cells, predict novel gene functions, and highlight critical genetic dependencies that may be exploited therapeutically to treat diseases such as cancer. We have used co-isogenic cell lines to map genetic interactions of selected metabolic query genes, which revealed novel insights into fatty acid metabolism and the functional role of a previously uncharacterized gene.
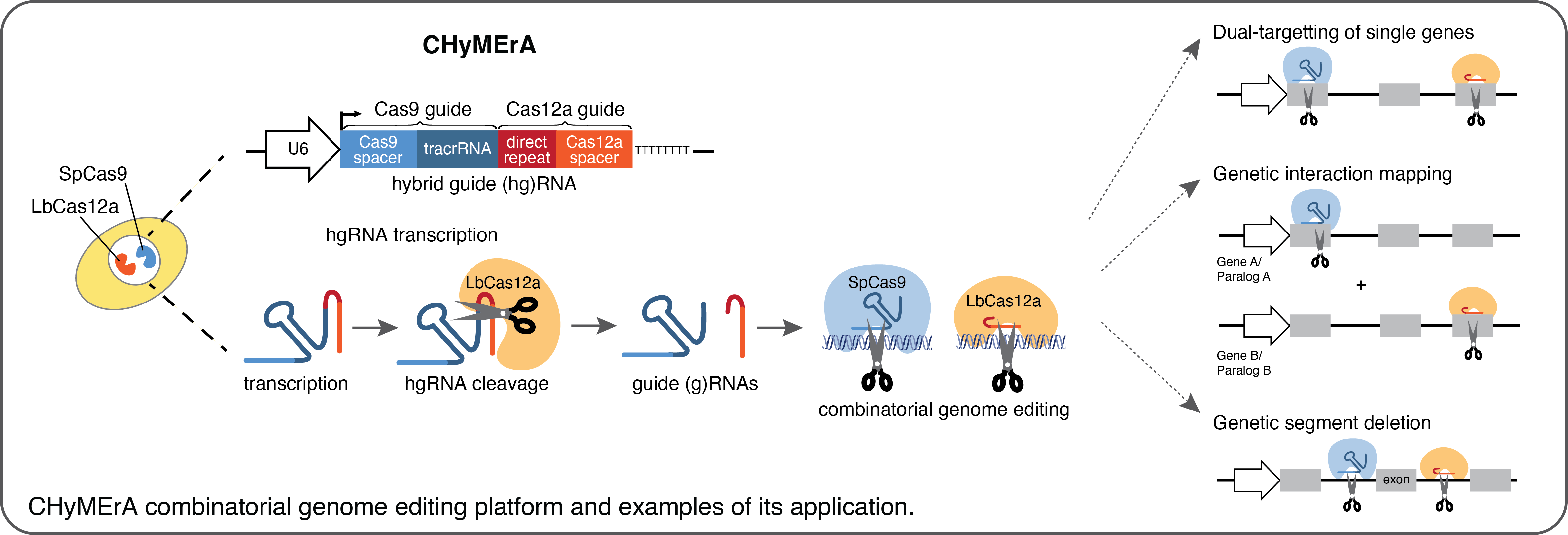
To expand our tools to map genetic interactions we developed CHyMErA (Cas Hybrid for Multiplexed Editing and Screening Applications), a CRISPR-based combinatorial screening platform for the perturbation of multiple genes. CHyMErA is based on the co-expression of Cas9 and Cas12a nucleases in conjunction with a hybrid guide RNA (hgRNA) engineered by the fusion of Cas9 and Cas12a guides and expressed from a single U6 promoter. We have applied CHyMErA for the dual-targeting of single genes, the mapping of genetic interactions between paralogs, and for exon-resolution functional genomics in mammalian cells. In our lab we continue exploring CHyMErA as a powerful tool to uncover genetic interactions and dependencies between various cellular pathways.
4. Technology development for systematic functional genomics approaches
We have a strong interest in developing and applying cutting-edge CRISPR-based technologies and continue exploring various Cas modalities and effector domains for our screening applications. Furthermore, we are also expanding the readouts for our screening platforms in order to assess genetic dependencies across more information-rich phenotypes.
Publications
Biography
Michael Aregger, Ph.D.
Dr. Michael Aregger obtained his Bachelor’s degree in Biology from the University of Basel, Switzerland in 2006 before earning his Master’s degree in 2008. In 2014, Dr. Aregger obtained his Ph.D. from the University of Dundee in Scotland where he worked in Prof. Victoria Cowling’s laboratory studying the regulation of mRNA cap methylation in human cells. For his postdoctoral research, he joined Prof. Jason Moffat’s laboratory at the University of Toronto in Canada to study genetic vulnerabilities and interactions in human cells. Dr. Aregger’s postdoctoral research has been recognized with multiple fellowships and awards, including the Donnelly Centre Research Excellence Award (2019) and two postdoctoral fellowships from the Swiss National Science Foundation (2014, 2016). In 2021, Dr. Aregger was recruited as a NIH Stadtman Investigator to the Molecular Targets Program at the National Cancer Institute to establish the Functional Genomics Section. His lab focuses on applying functional genomics approaches to explore genotype-to-phenotype relationships, gain insights into the plasticity of cancer cells and to identify genetic dependencies that may present novel therapeutic targets.
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Lab Life

Summer gathering of Aregger & Gonatopoulos-Pournatzis groups in Frederick celebrating Swati's completion of her PostBac training (06/2024).

The Aregger lab after having completed the scavenger hunt (06/2024)!
On the scavenger hunt through Downtown Frederick (06/2024).

Watching the partial solar eclipse outside our lab (04/2024).

Playing bowling during our 2023 holiday gathering (12/2023).

The Aregger Lab at the NIH RNA Day Picnic at Black Hill Regional Park (08/2023).

Summer trip to Harper's Ferry (WV) including a thrilling zipline ride through the trees (07/2023)!


Tyler in the air!

Saying goodbye to our PostBac students Tyler and Shreya...

Enjoying delicious Japanese food during our holiday gathering in Downton Frederick (12/2022).

Successs, we're happy to report that we managed to find Santa's list of gifts and escaped the Great Santascape room on time! Shreya proved herself as the master mind of the group while Chelsee showed great musical talent ;-)

Joint summer party with the Gonatopoulos-Pournatzis lab (07/2022).

At the same time we also wished farewell to our PostBac students Ethan and Belene as well as our summer student Anna.
Team
News
May 2025 - Congrats to Olga for winning two more Poster Prizes at the 2025 Frederick Spring Research Festival and the NIH PostBac Poster Day!
May 2025 - Chelsee and Olga each won a Presentation Award at the CCR Fellows & Young Investigators Colloquium for their talk and poster, respectively. Congrats to both!
May 2025 - Our recent exon-resolution functional genomics study is featured in the 2025 CCR Milestone magazine, which features CCR's top scientific advances from the past year!
November 2024 - Congrats to Youngkyu for winning a well-deserved Poster Award at the NCI RNA Biology Initiative Retreat 2024!
October 2024 - Our lab is growing. We are excited to welcome Ritam Naha as a new PostDoc to our team!
September 2024 - A very warm welcome to Olga Drozdovitch, who is joining our team as an iCURE PostBac student.
July 2024 - We say farewell to Swati who is starting grad school at Princeton University, NJ. Congrats on your awesome achievement and good luck with your PhD studies!
June 2024 - We're excited to report the publication of our lab's first paper in collaboration with the group of Thomas Gonatopoulos-Pournatzis. Check out our CRISPR genome-scale exon perturbation study published in Molecular Cell!
May 2024 - We welcome Ishaan Gollamudi who joins our lab for a NIH Summer Internship.
May 2024 - Swati's presentation of optimizing combinatorial CRISPRa systems was recognized with an Outstanding Poster Award at the NIH PostBac Poster Day 2024!
May 2024 - Chelsee has secured a 2024 CCR Health Disparities Travel Award, congrats!
April 2024 - Congratulations to Swati for winning a poster prize at the 2024 Spring Research Festival in Frederick!
November 2023 - Congrats to Chelsee who has been selected for a Sallie Rosen Kaplan (SRK) Postdoctoral Fellowship for Women Scientists!
October 2023 - Our team is further growing and we're excited to welcome Josef Horák as a new PostDoc to our group!
July 2023 - We say farewell to Tyler who is starting his MD studies at the University of Arizona in Tuscon, AZ. Congrats on your awesome achievement and good luck with Med School!
March 2023 - Youngkyu Jeon has joined our team as a PostDoc. We're excited to welcome Youngkyu at NCI-Frederick!
December 2022 - Welcome to Bandana Kumari who is joining our lab and the group of Dr. Gonatopoulos-Pournatzis as a computational PostDoc.
October 2022 - We're excited to further extend our team and to welcome Chelsee Holloway who is starting her PostDoc in our group.
September 2022 - Our group has been awarded a Pamela Anne Cafritz Renal Cell Carcinoma Award for the mapping of genetic interactions in patient-derived cancer models to uncover novel therapeutic targets for Renal Cell Carcinomas. We’re very grateful for the generous support of our research by the NCI's Center for Cancer Research and our collaborators Marston Linehan and Barry O'Keefe!
August 2022 - A warm welcome to Swati Sharma who joins our team as a PostBac student.
May 2022 - We welcome Anna Riordan who joins our group for a NIH Summer Internship.
May 2022 - Congratulations to Belene for winning a poster prize at the 2022 Virtual Postbac Poster Days!
October 2021 - The Aregger lab has officially opened. Welcome to the team Belene Oudit and Tyler On and thanks everyone for the warm welcome at the MTP and NCI-Frederick!