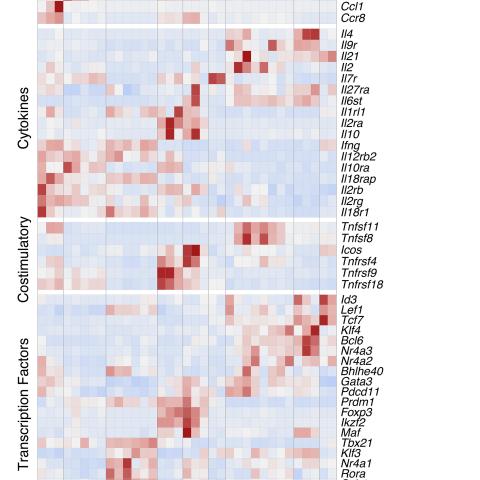
Transcriptomic Patterns of TILs, dLN Cells, and Arm Cells
Photo courtesy of Dr. Bosselut
Most cancer immunotherapy treatments harness powerful immune-system cells, known as CD8+ T cells, that can kill cancer cells that express a new tumor antigen (typically a neoantigen generated by a mutation within the cancer cell genome). CD4+ T cells, on the other hand, generally do not directly kill target cells but instead provide signals that help activate other immune cells. Because CD4+ T cells can elicit a powerful immune response, researchers have tried to harness such responses to fight cancer. However, the diversity of CD4+ T cells functions has so far been a serious obstacle to this endeavor.
A finding published in Cell Reports that holds promise for resolving some of the conundrums surrounding CD4+ T cells and their potential in fighting cancer comes from Rémy Bosselut, M.D., Ph.D., Senior Investigator in CCR’s Laboratory of Immune Cell Biology, and colleagues. This work, carried out in collaboration with Sridhar Hannenhali, Ph.D., who recently joined CCR from the University of Maryland, within the framework of a NCI-University of Maryland graduate partnership, found that CD4+ T-cell responses to tumor antigens are quite different from those to infections, highlighting a need to re-think how to harness the power of CD4+ T cells.
The researchers performed single-cell RNA sequencing on CD4+ T cells responding to an antigen expressed by mouse tumor cells. These CD4+ T cells were either from the tumor itself (tumor-infiltrating lymphocytes, or TILs) or from the lymph node draining the tumor (dLN T cells). TILs are immune cells that go from the bloodstream into a tumor to attack cancer cells; draining lymph nodes are important organs in which immune responses are typically initiated.
The findings identified multiple groups of tumor-responding CD4+ T cells, distinguished by gene expression (transcriptomic) patterns. Furthermore, transcriptomic patterns identified in TILs or dLN substantially differed from those seen in anti-viral CD4+ T cell responses.
Dr. Bosselut believes the patterns they’ve uncovered could form the basis for mining the transcriptome of human CD4+ TILs, with the long-term objective of developing indicators of a patient’s prognosis or optimizing treatment strategies. He notes, however, that these current findings need to be extended to other experimental systems and do not yet have any direct clinical application.
The investigators’ next steps will be to expand these findings to additional tumor models because a single model cannot probe the diversity of immune responses to tumors. They also hope to leverage their finding to analyze the CD4+ T cell response to human tumors as well as the functional relevance of the patterns they’ve detected.


