Daniël P. Melters, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 41, Room B1300
- Bethesda, MD 20892-5055
- 240-760-6664
- meltersdp@mail.nih.gov
RESEARCH SUMMARY
How are chromatin domains formed, how are they maintained, and how are they altered in cancer? The role of histone variants, in combination with nucleosome-binding proteins, orchestrate the functional aspects of chromatin domain, including regulating chromatin accessibility and transcriptional output. Using cell biological, biochemical, and biophysical tools, Dr. Melters dissects how chromatin accessibility is modulated by histone variants and chromatin-binding factors in cancer.
Areas of Expertise

Daniël P. Melters, Ph.D.
Publications
Biography

Daniël P. Melters, Ph.D.
Dr. Melters received his Bachelor's and Master’s degree in Biomedical Sciences from the Leiden University, the Netherlands. For his M.S .degree, he studied important questions in immunology and neuroscience. After his M.S., Dr. Melters joined the endocrinology lab of Dr. Pearce’s lab at UCSF. For his Ph.D., Dr. Melters joined the labs of Drs. Ian Korf and Simon Chan at UC Davis. In this collaboration, he used bioinformatic tools to identify candidate centromeric tandem repeats across the eukaryotic tree. In addition, he joined the biotechnology program, where he did an internship at Genentech in science policy. Dr. Melters joined the LRGBE/CSEM as a Postdoctoral Fellow, where he works under the guidance of Dr. Yamini Dalal, studying the structural features of centromeric chromatin in cancer, elucidating functional consequences when chromatin regulation goes awry in cancer.
Gallery
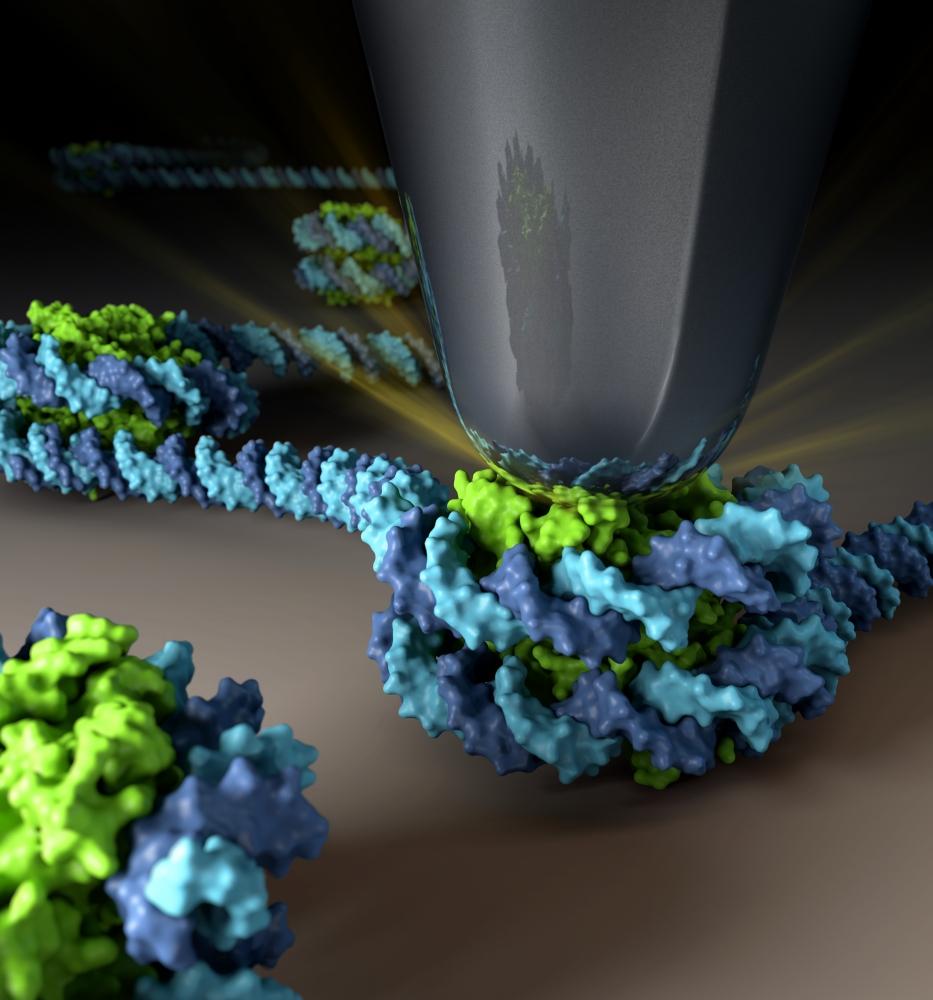
Atomic Force Microscope tip indenting a CENP-A nucleosome. Centromeric CENP-A nucleosomes are twice as elastic compared to canonical H3 nucleosomes and this material property can be modulated by the essential kinetochore protein CENP-C. Image made by NIH Medical Arts.
https://www.pnas.org/doi/10.1073/pnas.1911880116
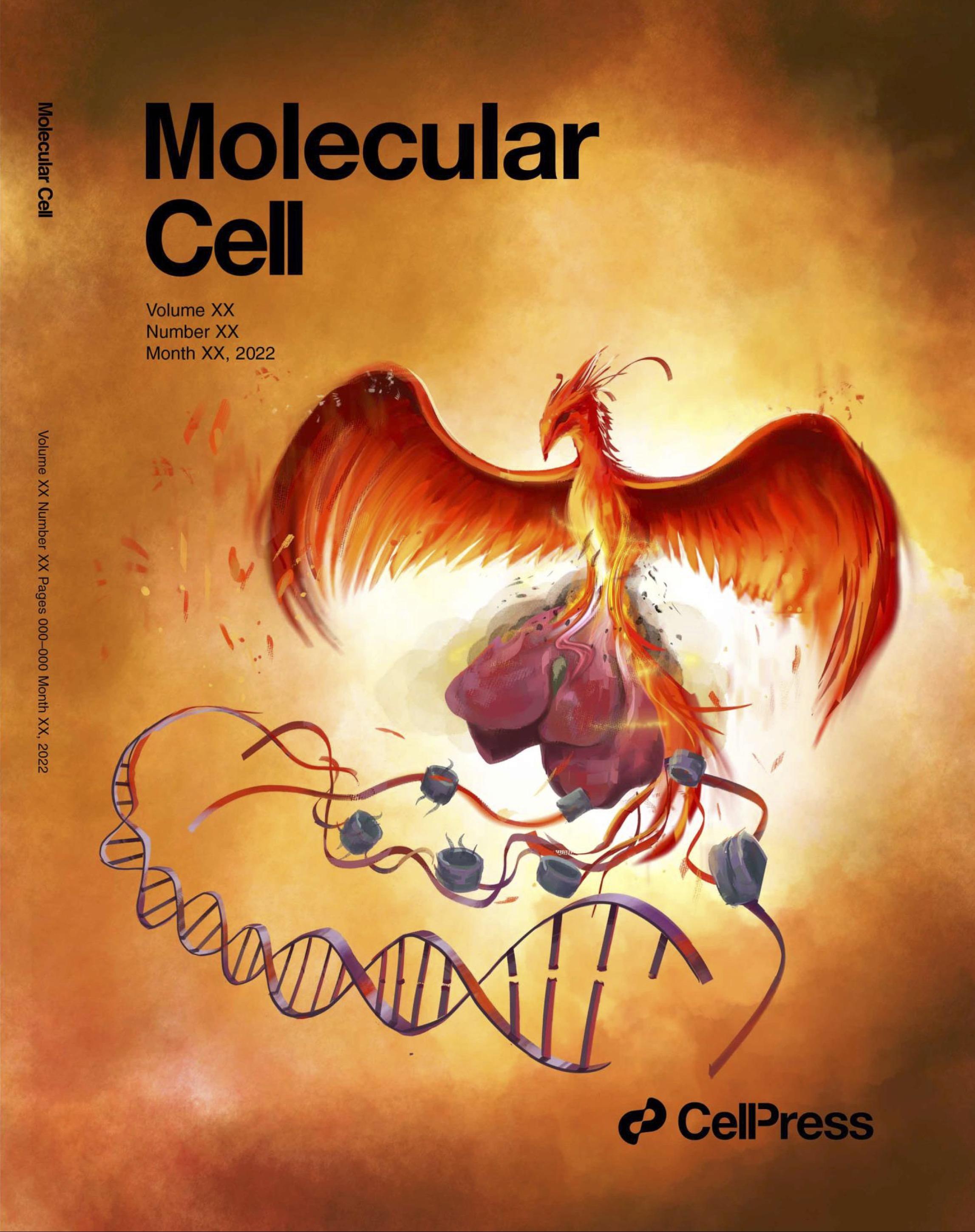
A phoenix offspring emerging from the ashes of an old liver during regeneration through chromatin-mediated mechanisms.
In this issue of Molecular Cell, Yang and colleagues discover age-dependent increases in broad regions of the repressive histone modification H3K27me3. They also demonstrate partial reversion to younger H3K27me3 patterns and gene expression upon resection of older livers.
Preview: https://www.sciencedirect.com/science/article/pii/S1097276523003180
Article: https://www.sciencedirect.com/science/article/pii/S1097276523002496?via%3Dihub

ON THE COVER
The essential kinetochore component CENP-C is critical in regulating centromere homeostasis. The image shows a multipolar spindle metaphase cell, a consequence when CENP-C is overexpressed. Mitotic spindles (cyan) attach to the chromosomes (grey) via the kinetochore (CENP-C in red).
https://www.life-science-alliance.org/content/8/4/e202402819