Xinhua Ji, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 538, Room 207
- Frederick, MD 21702
- 301-846-5035
- 301-846-6073
- jix@mail.nih.gov
RESEARCH SUMMARY
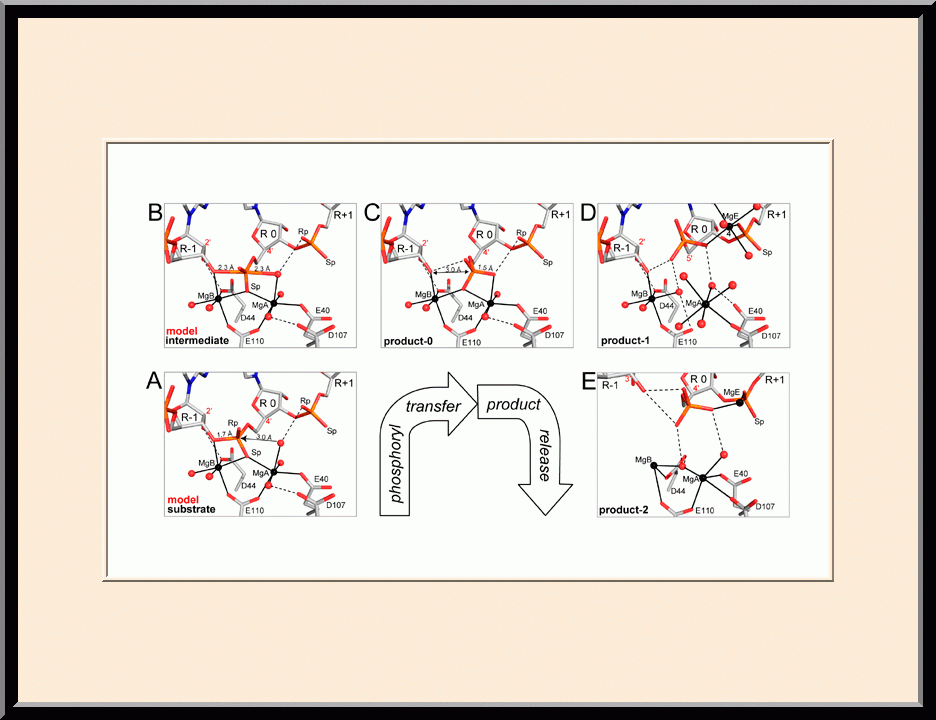
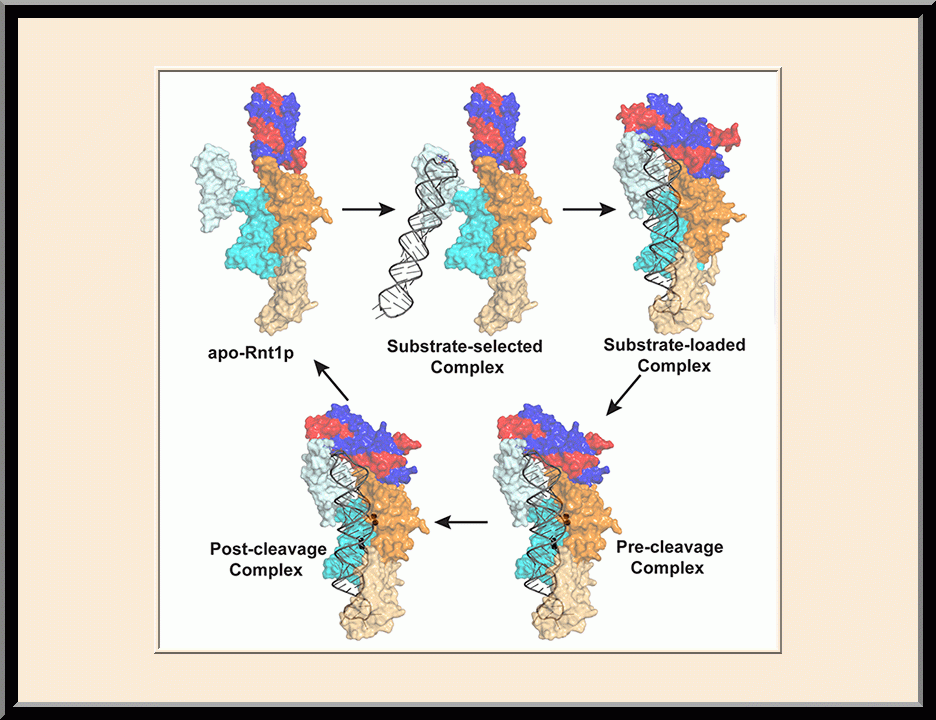
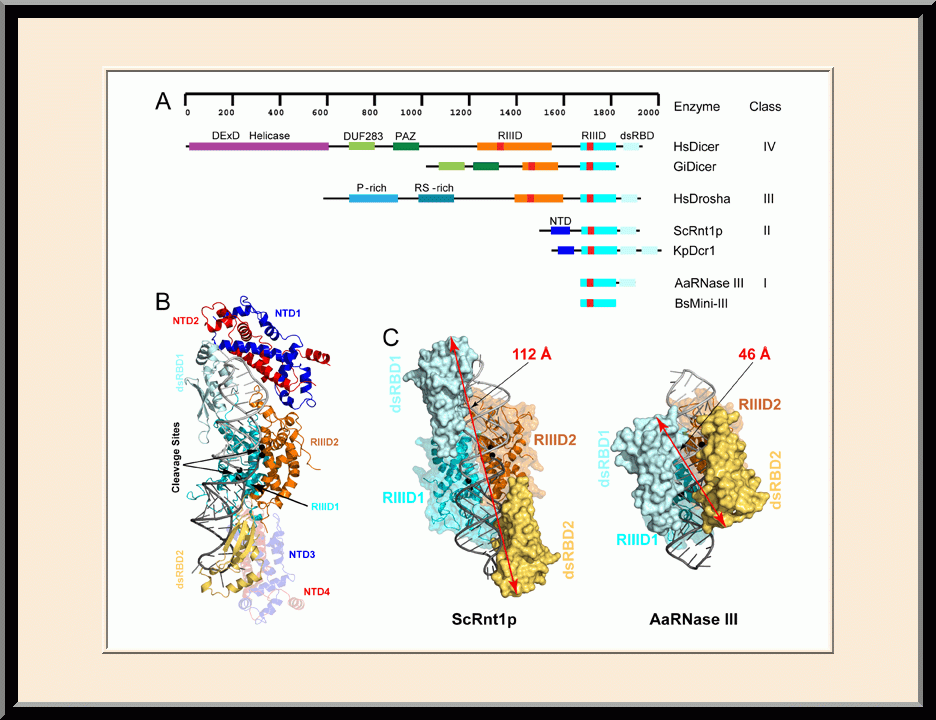
The major focus of Dr. Ji’s basic research is the structural biology of RNA biogenesis, with an emphasis on RNA-processing proteins, especially the ribonuclease III (RNase III) enzymes. He pioneered structural analysis of RNase IIIs in complex with double-stranded RNA (dsRNA) molecules. His bacterial RNase III:dsRNA structures provided the first structural view of dsRNA processing, revealing structural basis for substrate selection, RNA sequence specificity, and two-magnesium(II)-ion catalysis (PMID: 24274754). His yeast RNase III (Rnt1p):dsRNA structures established a double-ruler mechanism for substrate selection and provided further insights into RNA sequence specificity. Together, his structures showed that substrate selection by RNase IIIs is independent of cleavage, allowing the recognition of substrates with different structures while preserving the basic mechanism of cleavage, the two-magnesium(II)-ion catalysis (PMID: 30548404). His structures also revealed distinct features between prokaryotes and eukaryotes in the mechanism of two-magnesium(II)-ion catalysis. For the cleavage of each phosphodiester bond, prokaryotic catalysis involves a third magnesium(II) ion, whereas eukaryotic catalysis employs two more amino acid side chains (PMID: 35829618). Dr. Ji’s post-cleavage structures determined at a stage immediately after the cleavage of scissile bonds for both prokaryotic and eukaryotic RNase IIIs provide a framework for visualizing the cleavage assembly architecture of other RNase III enzymes, including prokaryotic Mini-III and eukaryotic Dcr1, Drosha, and Dicer.
Areas of Expertise

Xinhua Ji, Ph.D.
Research
Biomolecular Structure and Mechanism, Structure-Based Drug Design
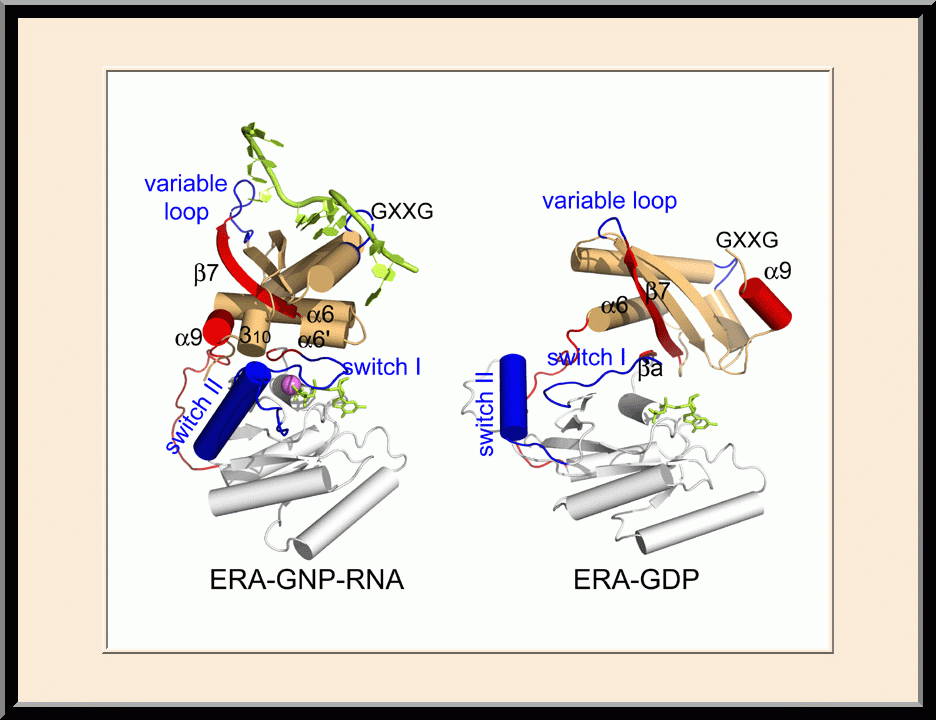
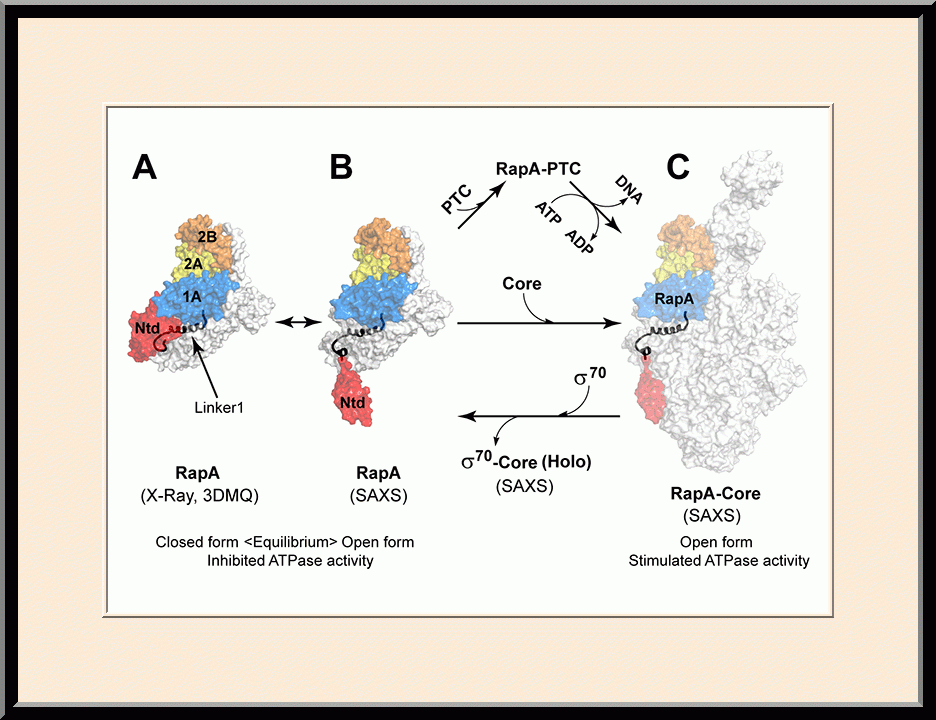
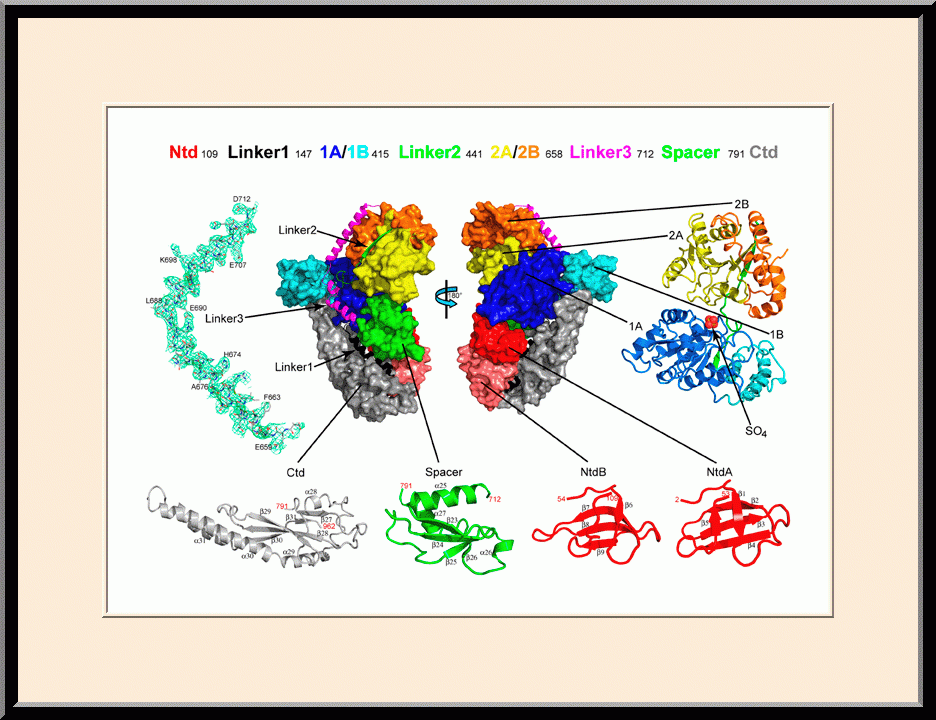
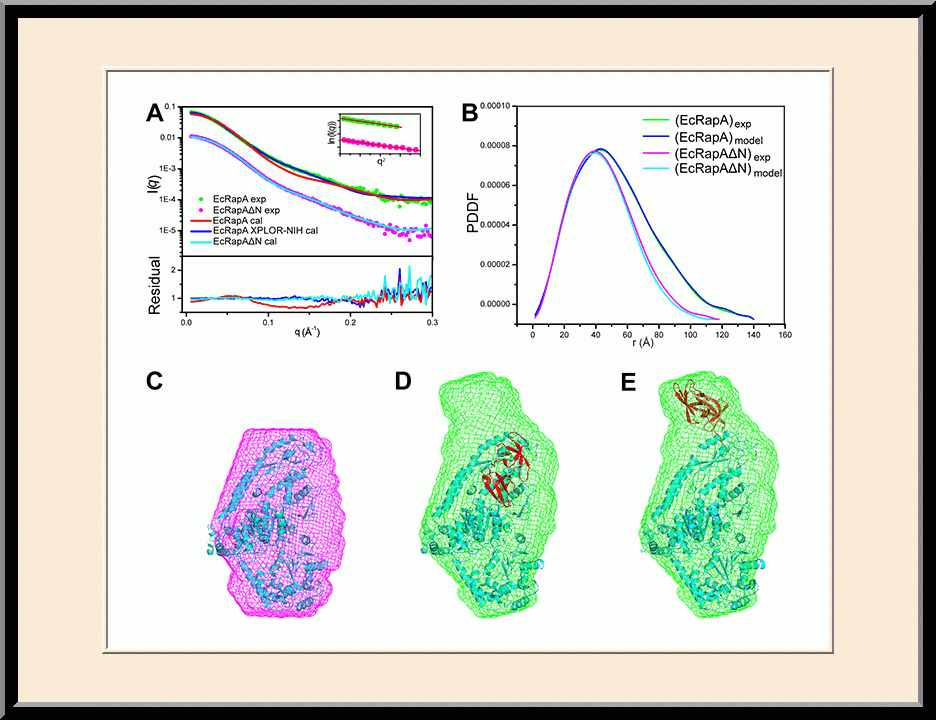
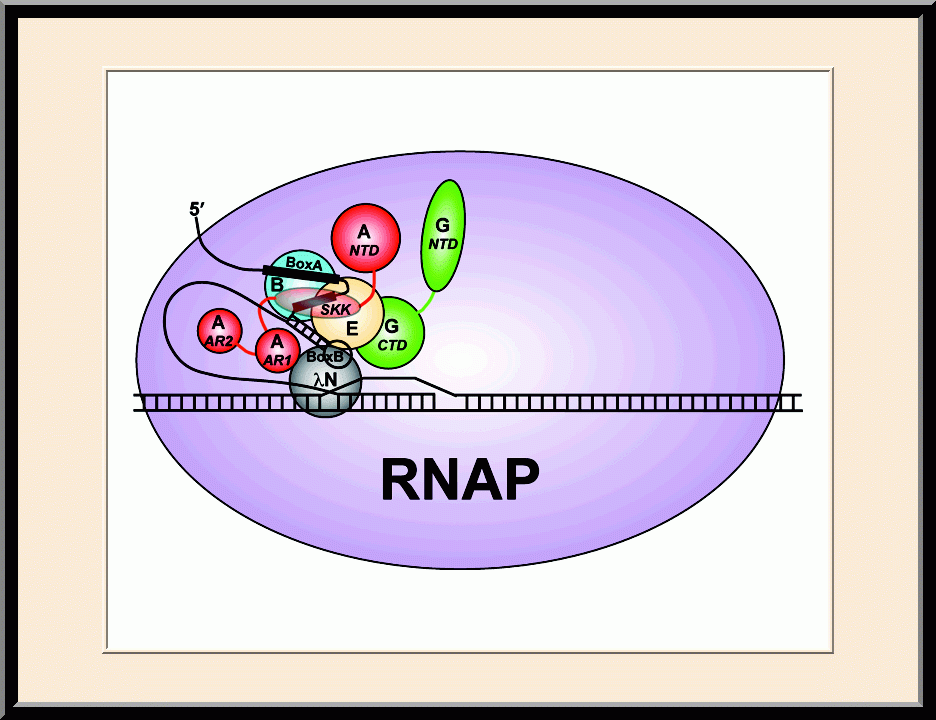
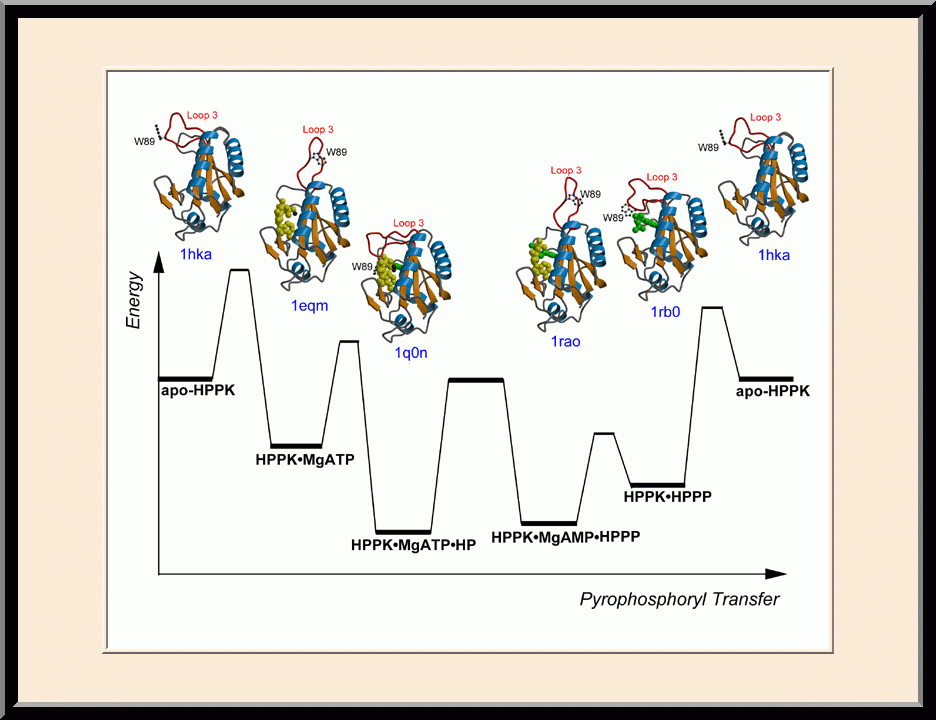
Our research is focused on the structural biology of RNA biogenesis, with an emphasis on RNA-processing proteins and RNA polymerase-associated transcription factors, and structure-based development of therapeutic agents. The goal of structural analysis is to map the reaction trajectory or functional cycle of selected biological macromolecules, and that of drug discovery is to design, synthesize, and characterize novel anticancer and antimicrobial agents. To date, we have described the reaction trajectory and/or functional cycle of HPPK (a folate pathway enzyme essential for microorganisms but absent in mammals), Era (an essential GTPase that couples cell growth with cell division), RapA (a Swi2/Snf2 protein that recycles RNA polymerase), and bacterial and yeast RNase III enzymes. Please see our Gallery for the structural view of these biological macromolecules and their reaction trajectory or functional cycle. Our contributions in RNase III research are summarized below, followed by examples of structure-based design of anticancer and antimicrobial agents.
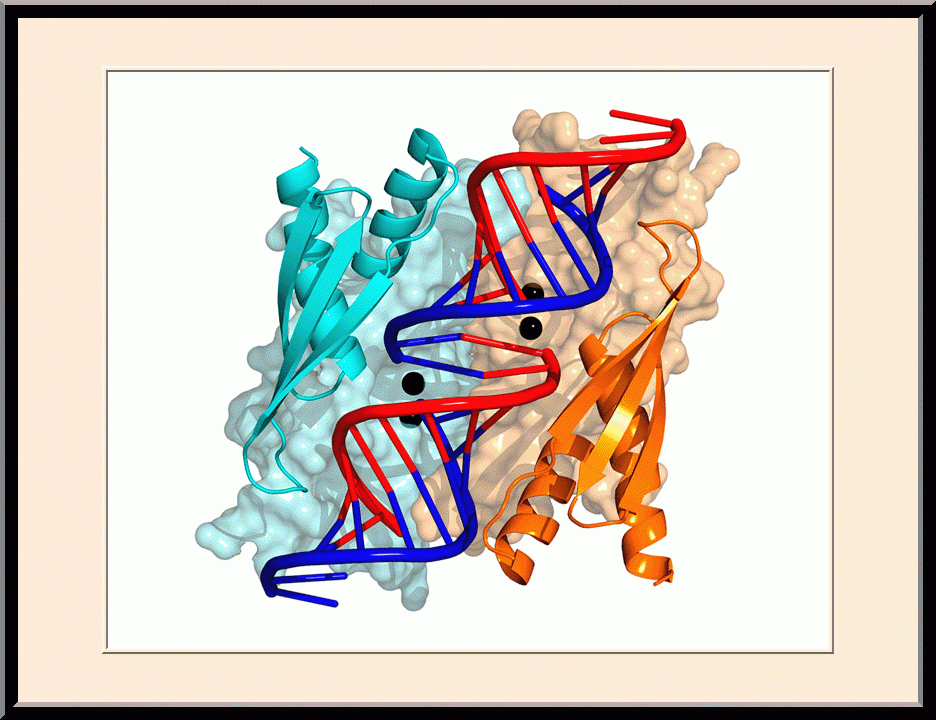
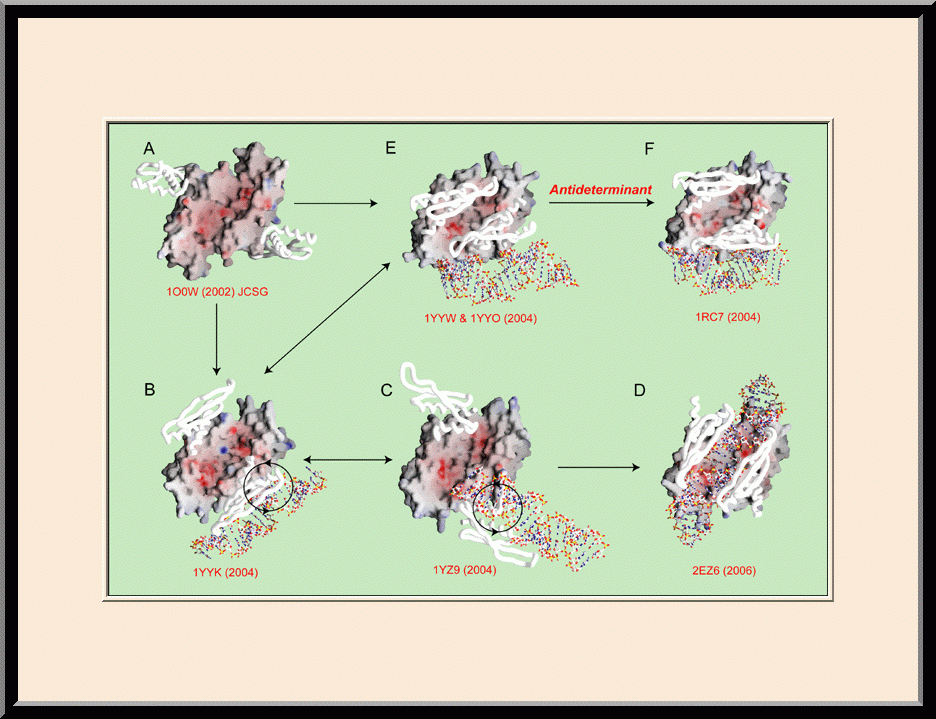
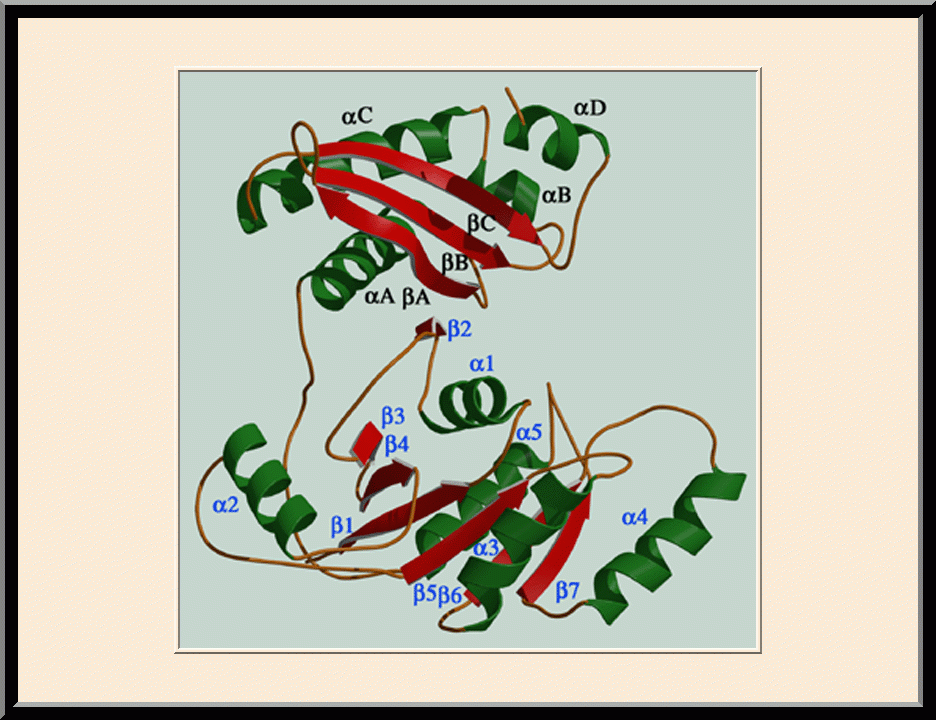
Structural view of RNA processing by RNase III enzymes. Representative members of the RNase III family include prokaryotic RNase III and eukaryotic Rnt1p, Drosha, and Dicer. They play important roles in RNA processing and maturation, post-transcriptional gene silencing, and defense against viral infection. For structural and mechanistic studies, bacterial RNase III and yeast Rnt1p are valuable model systems for prokaryotes and eukaryotes, respectively. For both RNase III and Rnt1p, we have shown how the dimerization of their endonuclease domain (RIIID) creates a catalytic valley where two cleavage sites are located, how the catalytic valley accommodates a dsRNA substrate in a manner such that each of the two RNA strands is aligned with one of the two cleavage sites, how the hydrolysis of each strand involves both RIIIDs, and how RNase III uses the two cleavage sites to create 2-nucleotide (2-nt) 3' overhangs in its products. We have also shown how magnesium is essential for the formation of a catalytically competent protein-RNA complex, and how the use of magnesium ions can drive the hydrolysis of each phosphodiester bond. Moreover, we have described a stepwise model by which RNase III and Rnt1p execute the phosphoryl transfer reaction. All members of the RNase III family propel RNA hydrolysis by two-Mg2+-ion catalysis, which exhibits distinct features, however, by prokaryotes and eukaryotes. As revealed by our structures, prokaryotic RNase IIIs require a third magnesium ion in catalysis, whereas eukaryotic RNase IIIs employ two additional amino acid side chains. For further details on bacterial RNase III and yeast Rnt1p, please see our review articles (PMID: 24274754; PMID: 30548404) and a recent report (PMID: 35829618).
Based on the protein-RNA interactions revealed by our structures of RNase III and Rnt1p, models with RNA can be meaningfully constructed for both Drosha and Dicer. A model complex of Drosha with RNA explains how Drosha enzymes recognize the last base pair in the basal junction of a primary microRNA substrate and measure 11 nucleotides up to position the scissile bond over the cleavage site. A model complex of Dicer with RNA explains how Dicer enzymes recognize the 2-nt 3' overhang of a dsRNA substrate and measure 22 nucleotides up to position the scissile bond over the cleavage site.
To fully characterize substrate specificity and product size of bacterial RNase III enzymes, we performed in vitro cleavage of dsRNAs by Ec and Aquifex aeolicus (Aa) RNase IIIs and delineated their products by next generation sequencing. Surprisingly, we found that both enzymes cleave dsRNA at preferred sites, among which a guanine nucleotide was enriched at a specific position (+3G). Based on sequence and structure analyses, we conclude that RNase IIIs recognize +3G via a conserved glutamine (EcQ165/AaQ161) side chain. Abolishing this interaction by mutating the glutamine to an alanine eliminates the observed +3G preference. Based on this finding, we created a research tool and named it bacterial Dicer that is ideally suited for producing heterogeneous siRNA cocktails to be used in gene silencing studies (PMID: 30916338).
Structure-based Development of Anticancer and Antimicrobial Agents. We carry out structure-based drug development primarily as a continuation of our basic research on the structure and mechanism of biomolecular systems with anticancer and antimicrobial significance. Previously, we designed PABA/NO, an enzymatically activated anticancer prodrug that kills cancer cells from within by releasing nitric oxide. We also made significant progress toward novel antibacterial agents targeting HPPK by the design and synthesis of linked purine pterin inhibitors and the determination of their crystal structures in complex with the enzyme. Recently, we have further developed both the PABA/NO and HPPK inhibitors. The PABA/NO derivative contains a PARP inhibitor and so it not only generates nitric oxide but also release a PARP inhibitor simultaneously in the same compartment, which damage DNA and inhibit its repair (PMID: 24521039; US Patent Number: 9168266). The improved HPPK inhibitors mimic closely the transition state of the catalytic complex and thereby have much higher potency (PMID: 33199204; US Patent Number: 11091509).
Publications
Structural basis for Dicer-like function of an engineered RNase III variant and insights into the reaction trajectory of two-Mg2+-ion catalysis
Structure and function of Rnt1p: An alternative to RNAi for targeted RNA degradation
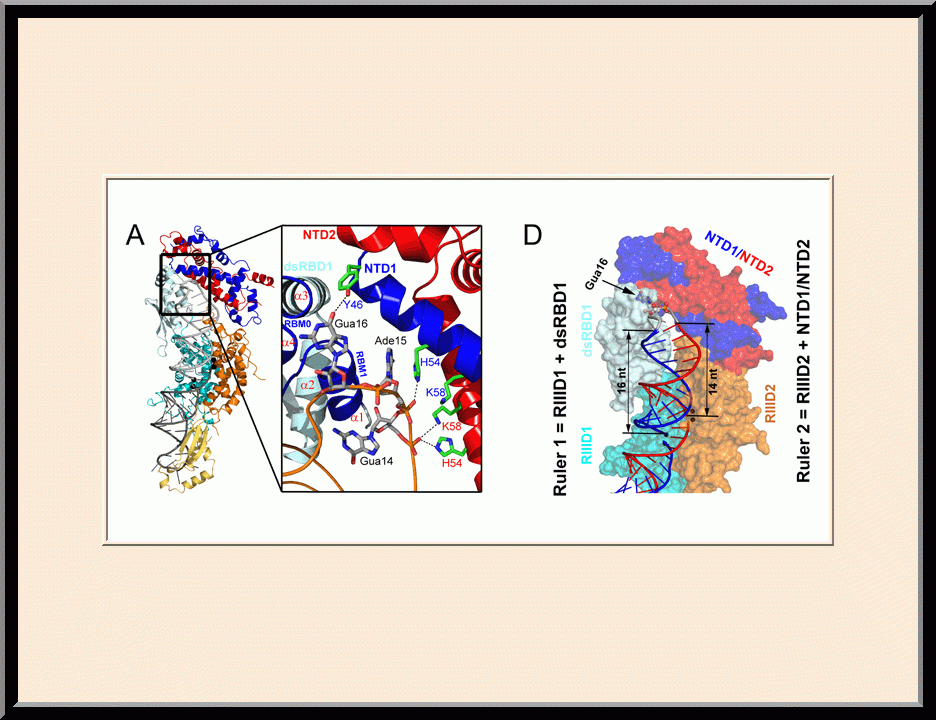
Structure of a Eukaryotic RNase III Postcleavage Complex Reveals a Double-Ruler Mechanism for Substrate Selection
RNase III: Genetics and Function; Structure and Mechanism
Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III
Biography

Xinhua Ji, Ph.D.
Dr. Ji earned his Ph.D. degree at the University of Oklahoma (1985-1990) and performed postdoctoral research at the University of Maryland (1991-1994), where he became a Research Assistant Professor before joining the National Cancer Institute (NCI), National Institutes of Health (NIH) in 1995. At the NCI at Frederick, Dr. Ji established his research directions (Biomolecular Structure and Mechanism, Structure-Based Drug Design) in the Advanced BioScience Laboratories (ABL)-Basic Research Program, moved to the Center for Cancer Research in 1999, and gained tenure in 2001 as an NIH Senior Investigator. He is a member of the American Crystallographic Association (1986-present), Chair/Co-chair of the NIH X-ray Diffraction Interest Group and Editor of its Newsletter (2001-2024), and a member of the Steering Committee of NCI RNA Biology Initiative (2018-2024).
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Team
News
- Winner of the NIH Fellows Award for Research Excellence (FARE) 2024 competition, “Cooperative unwinding of RNA duplex by DEAD-box helicase DDX3X, a new paradigm of division of labor,” Frederick, MD (2023).
- Invited speaker, the NIH RNA Club, "Structural basis for Dicer-like function of an engineered RNase III variant and insights into the reaction trajectory of two-Mg2+-ion catalysis," Bethesda, MD (2023).
- Recipient of the Federal Technology Transfer Award, "Structure-based development of HPPK inhibitors useful as antibacterial agents," Frederick, MD (2021).
- Invited speaker, the 17th Annual Congress of International Drug Discovery Science & Technology, “Structure-Based Design and Synthesis of HPPK Inhibitors," Kyoto, Japan (2019).
- Invited speaker, the International Conference on Translational Medicine, "Linked Purine Pterin HPPK Inhibitors Useful as Antibacterial Agents," San Antonio, Texas (2012).
- Recipient of the Federal Technology Transfer Award, "Structure-based development of HPPK inhibitors useful as antibacterial agents," Frederick, MD (2021).
- Invited speaker, the Shanxi Normal University, “Structure based drug design”, Xian, China (2013).
- Invited speaker, the Institute of Salt Lakes, Chinese Academy of Science, “Crystal structures of 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase and its reaction trajectory,” Xining, China (2007).
- Invited speaker, the Shanxi Normal University, “Crystal structures of Glutathione S-Transferase and its detoxification mechanism,” Xian, China (2004).
- Featured article on the front cover of the Journal of Bacteriology, “Overproduction of a dominant mutant of the conserved Era GTPase inhibits cell division in Escherichia coli," PMID: 32817092 (2020).
- Recipient of the Taylor & Francis Biomolecular Crystallography Poster Prize, the 70th Annual Meeting of the American Crystallographic Association, “Development of a model protein for HIV Tat structural study and drug development,” Virtual Meeting (2020).
- Representing a critical advance in coronavirus research, his article, “Structure of Severe Acute Respiratory Syndrome Coronavirus Receptor-binding Domain Complexed with Neutralizing Antibody" (PMID: 16597622), has been highlighted in a special virtue issue Coronaviruses (2020).
- Invited speaker, the III International Conference on Advanced Genetics, “The Molecular Mechanism of dsRNA Processing by a Bacterial Dicer,” Baltimore, MD (2019).
- Recipient of the Poster Award, the NCI Structural Biology Retreat, “The Molecular Mechanism of dsRNA Processing by a Bacterial Dicer,” Frederick, MD (2018).
- Invited speaker, the NIH RNA Club, "Characterization of dsRNA cleavage by the inside-out mechanism of ribonuclease III," Bethesda, MD (2016).
- Featured journal article on the front cover of MedChemComm, “Design, synthesis, and anticancer activity evaluation of irreversible allosteric inhibitors of ubiquitin-conjugating enzyme Ube2g2," PMID: 30542531 (2018).
- Invited speaker, the NIH RNA Club, “A Stepwise Model for Double-stranded RNA Processing by a Eukaryotic Ribonuclease III,” Bethesda, MD (2016).
- Invited speaker, the 2nd NCI Structural Biology Retreat, “RNA Recognition by hnRNP A1 and Its Regulatory Roles in MicroRNA Biogenesis,” Camp Greentop, Catoctin Mountain, MD (2016).
- Recipient of the Poster Award, the NCI Structural Biology Retreat, “Allosteric Activation of Bacterial Swi2/Snf2 Protein RapA by RNA Polymerase: Biochemical and Structural Studies," Frederick, MD (2015).
- Winner of the NIH Fellows Award for Research Excellence (FARE) 2016 competition, “Allosteric Activation of Bacterial Swi2/Snf2 Protein RapA by RNA Polymerase: Biochemical and Structural Studies,” Frederick, MD (2015).
- Featured journal article on the front cover of Molecular Cell, “Structure of a eukaryotic RNase III post-cleavage complex reveals a double-ruler mechanism for substrate selection," PMID: 24703949 (2014).
- Recipient of the Young Investigator Award, Young Investigator Travel Grant, and Finn World Travel Award, the 26th Annual Symposium of the Protein Society, “Structural Basis for Sequence Specificity and Product Length of Yeast Ribonuclease III,” San Diego, CA (2012).
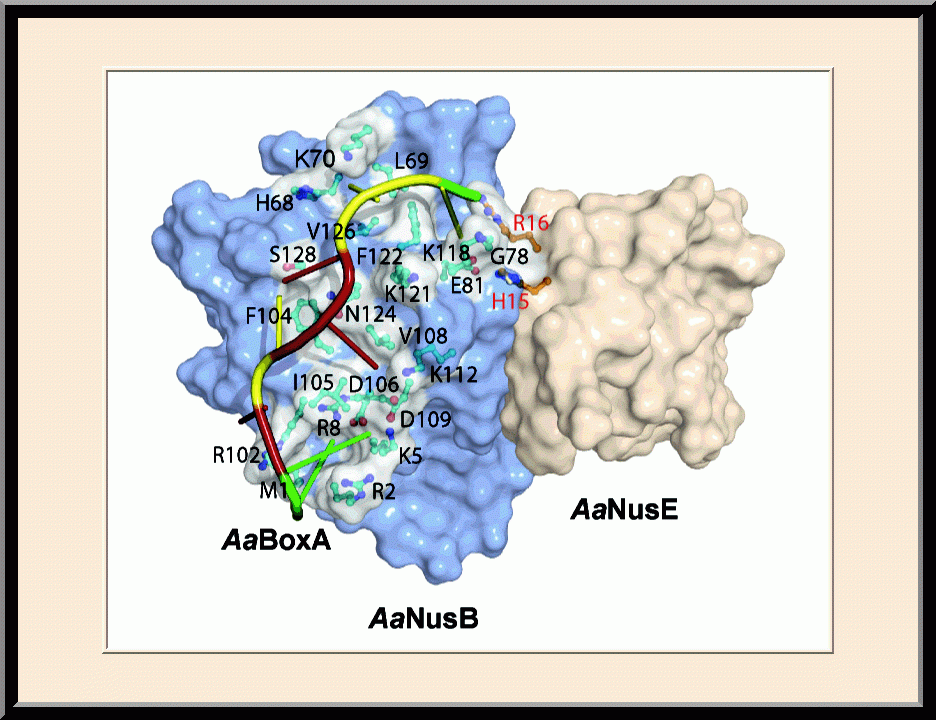
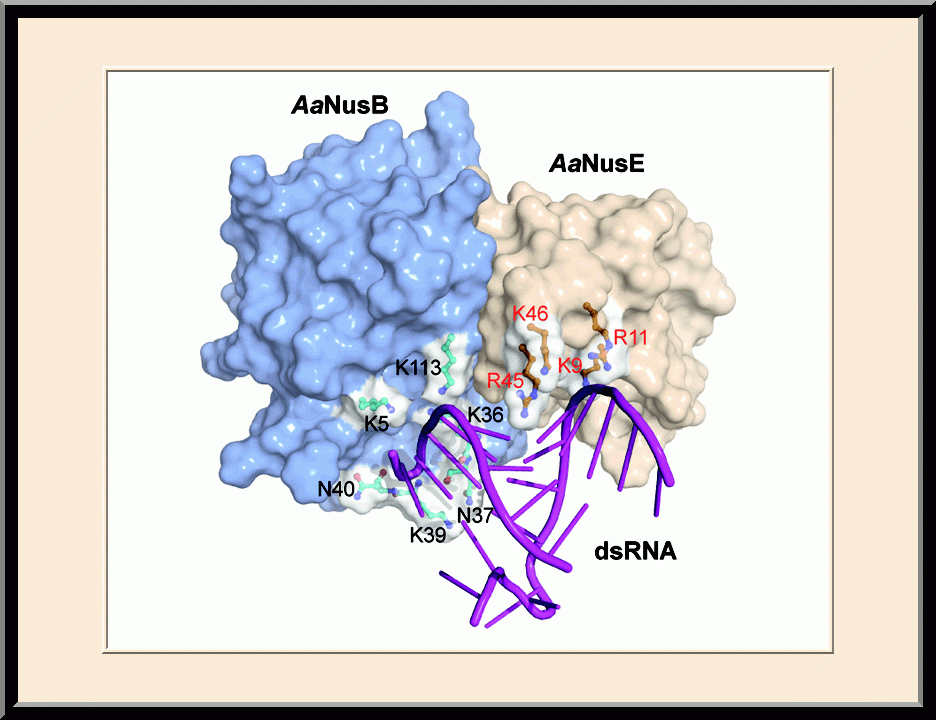
- Winner of the NIH Fellows Award for Research Excellence (FARE) 2013 competition, “Structural Basis for NusB in the Initiation of Transcription Antitermination and dsRNA Supercoiling,” Frederick, MD (2012).
- Recipient of the American Crystallographic Association Travel Award, the 62nd Annual Meeting of the American Crystallographic Association, “Crystal structure of a plectonemic RNA supercoil,” Boston, MA (2012).
- Recipient of the Young Investigator Award, the SER-CAT Annual Symposium, “Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination," Lexington, KY (2012).
- Invited speaker, the NIH Lambda Lunch Gala, “Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination,” Frederick, MD (2011).
- Invited speaker, the American Crystallographic Association Annual Meeting, “Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination,” New Orleans, LA (2011).
- Invited speaker, the NCI Structural Biophysics Laboratory, “Structural analysis of NusB-NusE-BoxA complexes reveals insight into antitermination complex assembly,” Frederick, MD (2010).
- Winner of the NIH Fellows Award for Research Excellence (FARE) 2010 competition, “Structure of ERA in complex with the 3′ end of 16S rRNA: Implications for ribosome biogenesis,” Frederick, MD (2009).
- Recipient of the Outstanding Scientific Presentation Award, the Spring Research Festival, “ERA: a GTP-dependent Molecular Switch Recognizes the 3’ End of 16S rRNA,” Frederick, MD (2009).
- Plenary speaker, the 17th Annual Microbial Genomics Conference, "Structure and dynamics of Era: a GTPase-dependent molecular switch recognizing the 3' end of 16S rRNA," Rocky Gap, MD (2009).
- Recipient of the Young Investigator Award, Young Investigator Travel Grant, and Finn World Travel Award, the 23rd Annual Symposium of the Protein Society, “Structure of ERA in complex with the 3′ end of 16S rRNA: Implications for ribosome biogenesis,” Boston, MA (2009).
- Recipient of the Poster Award, the Spring Research Festival, “The art of dsRNA processing by RNase III," Frederick, MD (2007).
- Recipient of the Young Investigator Award, the SER-CAT Annual Symposium, “Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III," Atlanta, GA (2006).
- Winner of the NIH Fellows Award for Research Excellence (FARE) 2007 competition, "The Mechanism of Double-Stranded RNA Cleavage by Ribonuclease III: How Dicer Dices,” Frederick, MD (2006).
- Textbook contribution: Figure 3 in her article, “Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents" (PMID: 10378268), has been reproduced in the college textbook BIOCHEMISTRY, starting from the 3rd edition, Donald Voet and Judith Voet (since 2002).
- Featured journal article on the front cover of Structure, “Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage," PMID: 11738048 (2001).
Covers

Overproduction of a Dominant Mutant of the Conserved Era GTPase Inhibits Cell Division in Escherichia coli
On the cover: Fluorescence microscopic image of Escherichia coli cells overproducing Era647, a dominant negative mutant mutant of the conserved GTPase Era. After several hours of Era647 overproduction, the cells cease dividing but continue to replicate and segregate their chromosomes (stained with 4ʹ,6-diamidino-2-phenylindole [DAPI], false-colored magenta). Nonfunctional aggregates of the cell division protein FtsZ (immunostained in green) localize exclusively to chromosome-free spaces in the cytoplasm. The inset shows one such cell, magnified x2, in which the magenta and green channels are offset to highlight the mutually specific localization of DNA and FtsZ under these conditions.
X. Zhou, H. Peters III, X. Li, N. Costantino, V. kumari, G. Shi, C. Tu, T. Cameron, D. Haeusser, D. Vega, X. Ji, W. Margolin, and D.L. Court, J. Bacteriol. 202, e00342-20 (2020). PMID: 32817092; PMCID: PMC7549363.

Design, synthesis, and anticancer activity evaluation of irreversible allosteric inhibitors of ubiquitin-conjugating enzyme Ube2g2
On the cover: The arrow represents an irreversible inhibitor, CW3. In the middle of the target is the thiol group of a cystine side chain at one edge of CW3-binding site in a ubiquitin-conjugating enzyme (E2), Ube2g2, the cognate E2 of the RING finger-dependent ubiquitin ligase (E3) gp78. Known as the tumor autocrine motility factor receptor, gp78 contributes to tumor progression. It interacts with Ube2g2 via two structural motifs in a stepwise, allosteric fashion. Forming a covalent bond with the cystine side chain, CW3 irreversibly blocks such allosteric interactions and thereby eliminates the tumorigenic effect of gp78. Among 19 compounds screened with the NCI 60 tumor cell lines, CW3 exhibited outstanding anticancer activities. At 10 μM, it caused >50% growth inhibition to 40% of the cell lines; at 100 μM, it showed lethiferous activity against most cell lines.
C. Wang, G. Shi, and X. Ji, MedChemComm 9, 1771-1974 (2018). PMID: 30542531; PMCID: PMC6238722.

Structure of a Eukaryotic RNase III Postcleavage Complex Reveals a Double- Ruler Mechanism for Substrate Selection
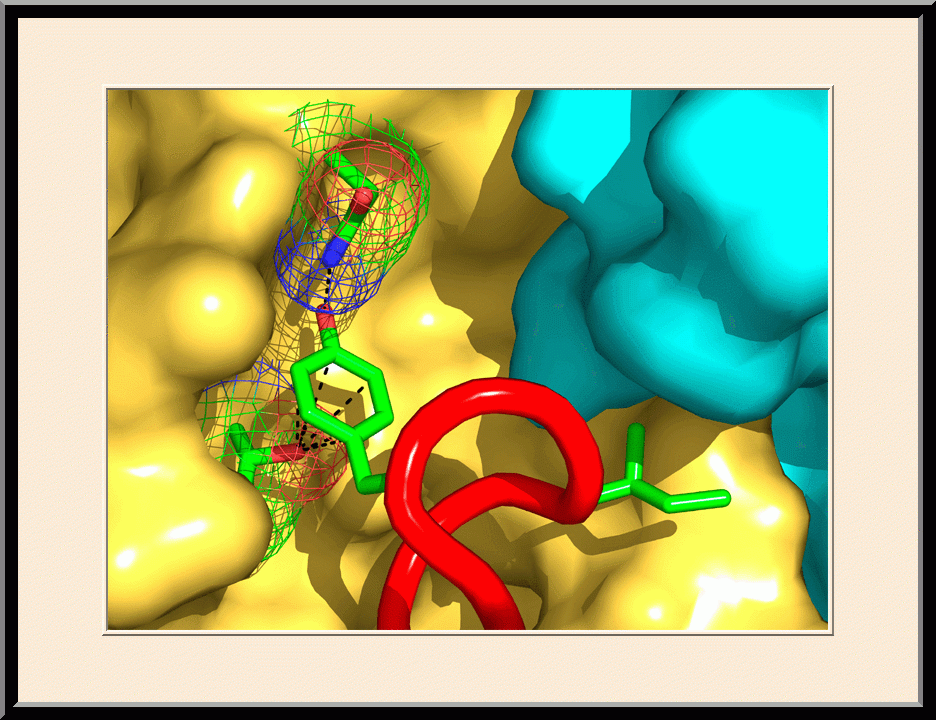
On the cover: The yeast RNase III (Rnt1p) selectively processes stem-loop RNAs with a guanine nucleotide in the second position of the capping tetraloop. In this issue, Liang et al. (pp. 431-444) show that Rnt1p recognizes the guanine nucleotide by a G clamp motif at the C terminus of dsRBD, that the guanine base is also recognized by the NTD, and that the G clamp is required for substrate binding and cleavage, whereas the NTD is required for the accuracy of processing. The cover illustrates guanine-specific substrate identification by the G clamp motif of Rnt1p as the cherry-picking hand that selectively harvests guanine-containing fruits. The hand, fingers, and cherries represent Rnt1p, G clamp, and different nucleotides in the second position of the tetraloop, respectively.
Y.-H. Liang, M. Lavoie, M.-A. Comeau, S. Abou Elela, and X. Ji, Mol. Cell 54, 431-444 (2014). PMID: 24703949; PMCID: PMC4019767.
Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage
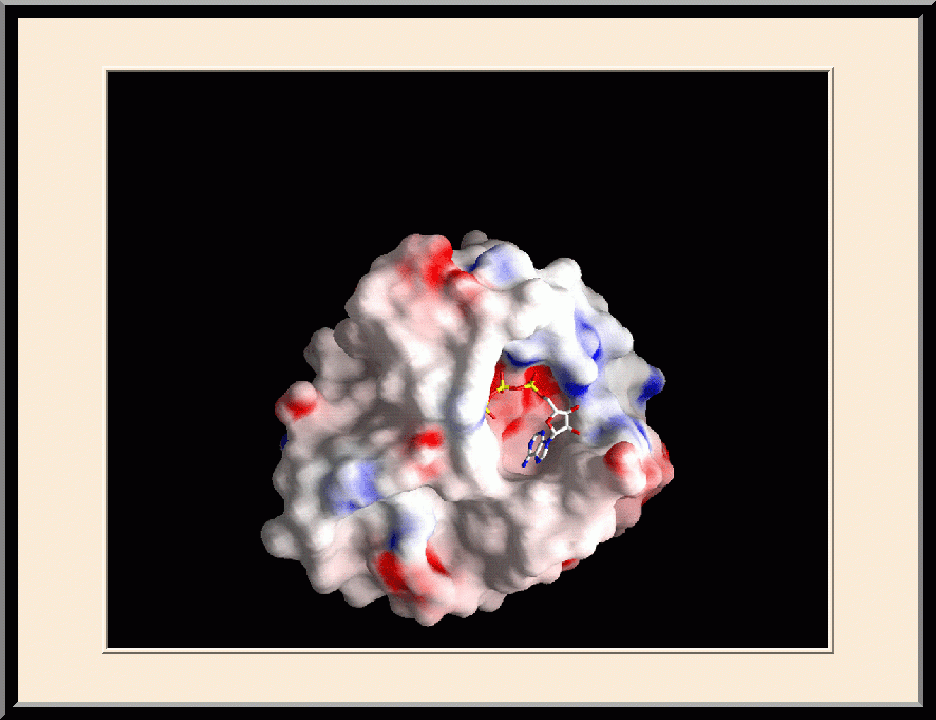
On the Cover: Model of a bacterial ribonuclease III (RNase III) in complex with a product of double-stranded RNA cleavage. The protein is shown as a surface representation with positive potential indicated in blue and negative in red. The product, shown as a stick model in green, has a total length of 13 nucleotides (9+2+2) created by two overlapping strands of 11 nucleotides each. The typical 2-nucleotide 3' overhang is seen at each end of the product.
J. Blaszczyk, J. E. Tropea, K. M. Routzahn, D. S. Waugh, M. Bubunenko, D. L. Court, and X. Ji, Structure 9, 1225-1236 (2001). PMID: 11738048.