Mary N. Carrington, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 37, Room 3032A
- Bethesda, MD 20814
- 240-760-6055
- carringm@mail.nih.gov
RESEARCH SUMMARY
Dr. Carrington is a senior principal scientist at the Frederick National Laboratory for Cancer Research and head of the HLA Immunogenetics Section in the Laboratory of Integrative Cancer Immunology Program. Her group studies the influence of genetic variation at loci encoding the human leukocyte antigen (HLA), killer cell immunoglobulin-like receptors (KIR), and other immune-related molecules on risk of and outcomes to infection, cancer, autoimmunity, and maternal-fetal disease. Recent studies have focused on identification of HLA allele-specific properties that affect their function beyond peptide binding specificity, such as differential gene expression, dependence on proteins of the antigen processing pathway, and interactions with specific KIR allotypes. Once values are assigned to these allele-specific attributes, they are then assessed for their impact on disease outcome.
Areas of Expertise
Mary N. Carrington, Ph.D.
Research
The main goal of the Carrington laboratory is to understand the genetic basis for resistance or susceptibility to human disease conferred by polymorphic immune response loci, including human leukocyte antigen (HLA), killer immunoglobulin-like receptor (KIR) genes and others. We study a large variety of disorders including cancer, transplantation outcome, autoimmune and infectious diseases. Our multidisciplinary team consists of experimental biologists and computational scientists.
Extensive genetic polymorphism is a primary characteristic of the HLA class I and class II loci, which encode cell surface molecules that present antigenic peptides to T cells, initiating an adaptive immune response. Variation within these loci is concentrated at positions that determine specificity for peptides, which can affect efficiency of immune responses. Thus, allelic composition of a given genotype determines the diversity of the peptide repertoire presented by a carrier of that genotype. Using mass-spectrometry data, we have been developing analytical tools to estimate the breadth of peptide repertoire based on HLA genotype.
Apart from determining antigenic peptide specificity, polymorphism at the HLA loci can differentially impact various functional properties of HLA molecules. These include expression levels, dependence on the intracellular chaperone tapasin for peptide loading, and interactions with immune receptors, such as KIR and leukocyte immunoglobulin-like receptors (LILR) expressed on the surface of innate immune cells. Using experimental data, we have developed models to test these allele-specific properties for their influence on disease outcomes. Our lab has shown that these genetically defined HLA features associate with HIV-1 pathogenesis, outcome to hematopoietic transplantion, and other diseases.
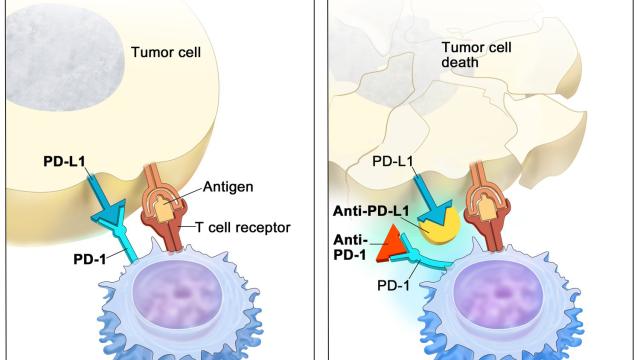
Variation at the HLA loci has been a major focus in genetic studies of cancer immunotherapy outcomes. We performed the largest epidemiological study of HLA class I alleles in cancer immunotherapy patients to date, and discovered that HLA-A*03 is associated with poor response to immunotherapy, which can be now used as a predictive marker.
Recent emphasis in our lab has focused on the non-classical HLA class I molecule HLA-E, which regulates immune responses through interactions with the inhibitory CD94-NKG2A and activating CD94-NKG2C receptors expressed by subsets of NK and T cells. HLA-E binds peptides derived from the signal sequences of the classical HLA-A, HLA-B, and HLA-C, and polymorphism in the HLA class I signal sequences differentially influence both HLA-E expression levels and interactions with CD94-NKG2A/C receptors. We are systematically investigating the impact of signal sequence polymorphism on HLA-E function using in vitro models and ex vivo analysis of NK cells. Our aim is to build a prediction algorithm to estimate the strength of HLA-E-mediated regulation of immune responses based on HLA class I genotype. Given the ability of HLA-E to regulate anti-viral and anti-tumor immune responses, these studies may inform vaccine and therapy design.
In the era where massive datasets are publicly available, and our own immunogenetic database is extensive, we are particularly interested in developing novel computational approaches that will take advantage of these resources. These tools will allow us to estimate the effects of variation in immune response genes on human disease by combining the knowledge of genetic association data with biological function, and to this end we are involved in analyses of several large biobank-based studies.
Publications
Genetic variation that determines TAPBP expression levels associates with the course of malaria in an HLA allotype-dependent manner
HLA-A*03 and response to immune checkpoint blockade in cancer: an epidemiological biomarker study
HLA tapasin independence: broader peptide repertoire and HIV control
The Role of HLA-DP Expression in Graft-Versus-Host Disease After Unrelated Donor Transplantation
Biography
Mary N. Carrington, Ph.D.
Dr. Carrington obtained her Ph.D. at Iowa State University in the Immunobiology Department. She performed her postdoctoral studies in the departments of Immunology and Microbiology at Duke University and the University of North Carolina. Prior to her current appointment, she was a faculty member in the Immunology Department at Duke University. Dr. Carrington serves as a Steering Committee member of the Ragon Institute of MGH, MIT, and Harvard. In 2022, she was elected to the American Academy of Arts and Sciences.
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Team
News
Renowned FNL scientist joins late father-in-law and world leaders as member of American Academy of Arts and Sciences
Earlier this year, Dr. Mary Carrington was elected to the prestigious American Academy of Arts and Sciences. She will be officially inducted sometime in 2023. https://frederick.cancer.gov/news/renowned-fnl-scientist-joins-late-father-law-and-world-leaders-member
Understanding the Foundations of Immune Defenses
Dr. Mary Carrington elected to the American Academy of Arts and Sciences for Insights into Immune System Variations. https://irp.nih.gov/blog/post/2022/08/understanding-the-foundations-of-immune-defenses
Oyez, Oyez, Oyez! Saunders PM, Brooks AG, Rossjohn J.Nat Immunol. 2023 Jul;24(7):1052-1053. doi: 10.1038/s41590-023-01541-x.PMID: 37308666